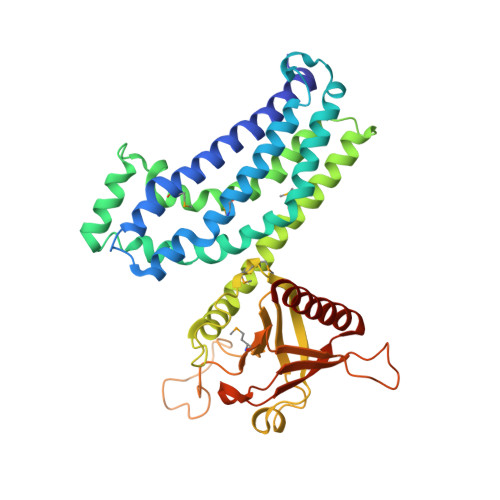
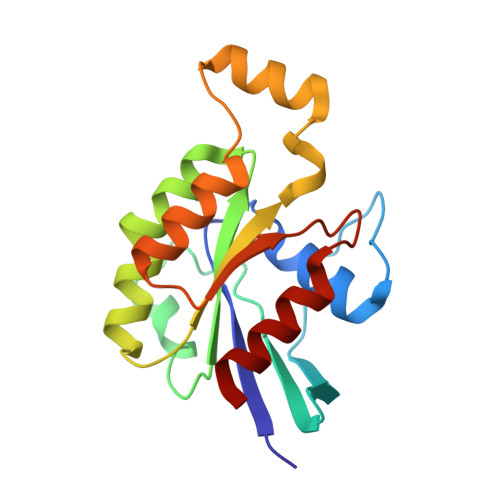
Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1.
Worthylake, D.K., Rossman, K.L., Sondek, J.(2000) Nature 408: 682-688
- PubMed: 11130063
- DOI: https://doi.org/10.1038/35047014
- Primary Citation of Related Structures:
1FOE - PubMed Abstract:
The principal guanine nucleotide exchange factors for Rho family G proteins contain tandem Dbl-homology (DH) and pleckstrin-homology (PH) domains that catalyse nucleotide exchange and the activation of G proteins. Here we have determined the crystal structure of the DH and PH domains of the T-lymphoma invasion and metastasis factor 1 (Tiam1) protein in complex with its cognate Rho family G protein, Rac1. The two switch regions of Rac1 are stabilized in conformations that disrupt both magnesium binding and guanine nucleotide interaction. The resulting cleft in Rac1 is devoid of nucleotide and highly exposed to solvent. The PH domain of Tiam1 does not contact Rac1, and the position and orientation of the PH domain is markedly altered relative to the structure of the uncomplexed, GTPase-free DH/PH element from Sos1. The Tiam1/Rac1 structure highlights the interactions that catalyse nucleotide exchange on Rho family G proteins, and illustrates structural determinants dictating specificity between individual Rho family members and their associated Dbl-related guanine nucleotide exchange factors.
Organizational Affiliation:
Department of Pharmacology, Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, 27599, USA.