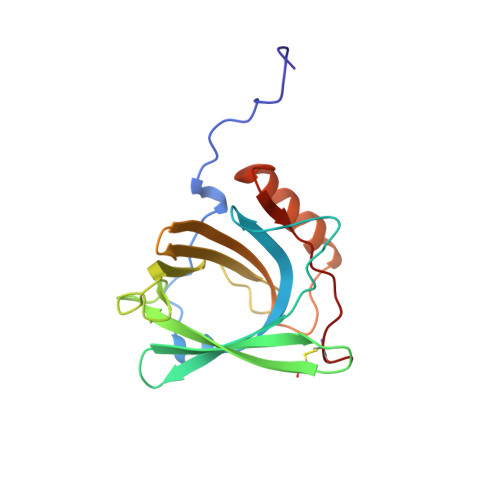
The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin.
Coles, M., Diercks, T., Muehlenweg, B., Bartsch, S., Zolzer, V., Tschesche, H., Kessler, H.(1999) J Mol Biol 289: 139-157
- PubMed: 10339412
- DOI: https://doi.org/10.1006/jmbi.1999.2755
- Primary Citation of Related Structures:
1NGL - PubMed Abstract:
Human neutrophil gelatinase-associated lipocalin (HNGAL) is a member of the lipocalin family of extracellular proteins that function as transporters of small, hydrophobic molecules. HNGAL, a component of human blood granulocytes, binds bacterially derived formyl peptides that act as chemotactic agents and induce leukocyte granule discharge. HNGAL also forms a complex with the proenzyme form of matrix metalloproteinase-9 (pro-MMP-9, or progelatinase B) via an intermolecular disulphide bridge. This association allows the subsequent formation of ternary and quaternary metalloproteinase/inhibitor complexes that vary greatly in their metalloproteinase activities. The structure and dynamics of apo-HNGAL have been determined by NMR spectroscopy. Simulated annealing calculations yielded a set of 20 convergent structures with an average backbone RMSD from mean coordinate positions of 0. 79(+/-0.13) A over secondary structure elements. The overall rotational correlation time (13.3 ns) derived from15N relaxation data is consistent with a monomeric protein of the size of HNGAL (179 residues) under the experimental conditions (1.4 mM protein, pH 6.0, 24.5 degrees C). The structure features an eight-stranded antiparallel beta-barrel, typical of the lipocalin family. One end of the barrel is open, providing access to the binding site within the barrel cavity, while the other is closed by a short 310-helix. The free cysteine residue required for association with pro-MMP-9 lies in an inter-strand loop at the closed end of the barrel. The structure provides a detailed model of the ligand-binding site and has led to the proposal of a site for pro-MMP-9 association. Dynamic data correlate well with structural features, which has allowed us to investigate a mechanism by which a cell-surface receptor might distinguish between apo and holo-HNGAL through conformational changes at the open end of the barrel.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Technische Universität München, Lichtenbergstrasse 4, Garching, 85747, Germany.