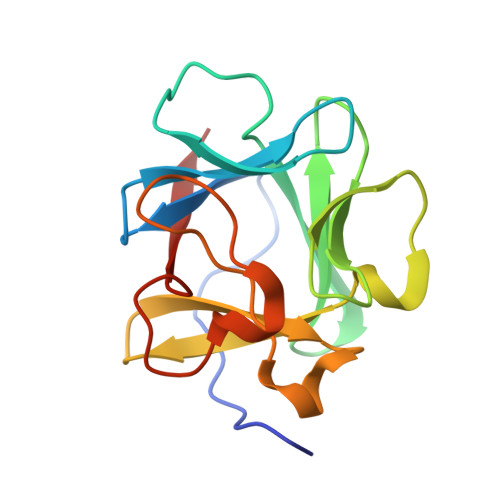
An atomic resolution structure for human fibroblast growth factor 1.
Bernett, M.J., Somasundaram, T., Blaber, M.(2004) Proteins 57: 626-634
- PubMed: 15382229
- DOI: https://doi.org/10.1002/prot.20239
- Primary Citation of Related Structures:
1RG8 - PubMed Abstract:
A 1.10-A atomic resolution X-ray structure of human fibroblast growth factor 1 (FGF-1), a member of the beta-trefoil superfold, has been determined. The beta-trefoil is one of 10 fundamental protein superfolds and is the only superfold to exhibit 3-fold structural symmetry (comprising 3 "trefoil" units). The quality of the diffraction data permits unambiguous assignment of Asn, Gln, and His rotamers, Pro ring pucker, as well as refinement of atomic anisotropic displacement parameters (ADPs). The FGF-1 structure exhibits numerous core-packing defects, detectable using a 1.0-A probe radius. In addition to contributing to the relatively low thermal stability of FGF-1, these defects may also permit domain motions within the structure. The availability of refined ADPs allows a translation/libration/screw (TLS) analysis of putative rigid body domains. The TLS analysis shows that beta-strands 6-12 together form a rigid body, and there is a clear demarcation in TLS motions between the adjacent carboxyl- and amino-termini. Although separate from beta-strands 6-12, the individual beta-strands 1-5 do not exhibit correlated motions; thus, this region appears to be comparatively flexible. The heparin-binding contacts of FGF-1 are located within beta-strands 6-12; conversely, a significant portion of the receptor-binding contacts are located within beta-strands 1-5. Thus, the observed rigid body motion in FGF-1 appears related to the ligand-binding functionalities.
Organizational Affiliation:
Institute of Molecular Biophysics and Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306-4380, USA.