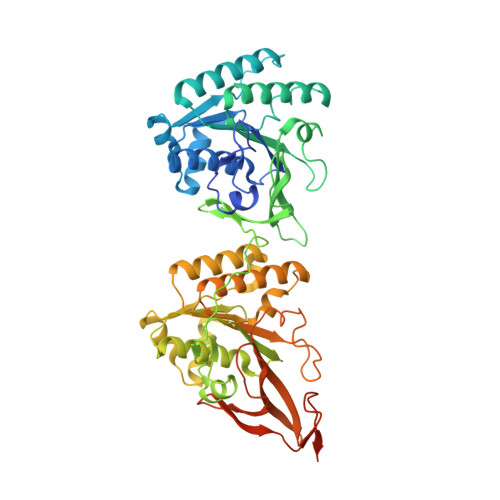
Visualizing a Circadian Clock Protein; Crystal Structure of KaiC and Functional Insights
Pattanayek, R., Wang, J., Mori, T., Xu, Y., Johnson, C.H., Egli, M.(2004) Mol Cell 15: 375-388
- PubMed: 15304218
- DOI: https://doi.org/10.1016/j.molcel.2004.07.013
- Primary Citation of Related Structures:
1TF7 - PubMed Abstract:
Circadian (daily) biological clocks express characteristics that are difficult to explain by known biochemical mechanisms, and will ultimately require characterizing the structures, functions, and interactions of their molecular components. KaiC is an essential circadian protein in cyanobacteria that forms the core of the KaiABC clock protein complex. We report the crystal structure of the KaiC homohexameric complex at 2.8 A resolution. The structure resembles a double doughnut with a central pore that is partially sealed at one end. The crystal structure reveals ATP binding, inter-subunit organization, a scaffold for Kai-protein complex formation, the location of critical KaiC mutations, and evolutionary relationships to other proteins. A key auto-phosphorylation site on KaiC (T432) is identified from the crystal structure, and mutation of this residue abolishes circadian rhythmicity. The crystal structure of KaiC will be essential for understanding this circadian clockwork and for establishing its links to global gene expression.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University, Nashville, TN 37232, USA.