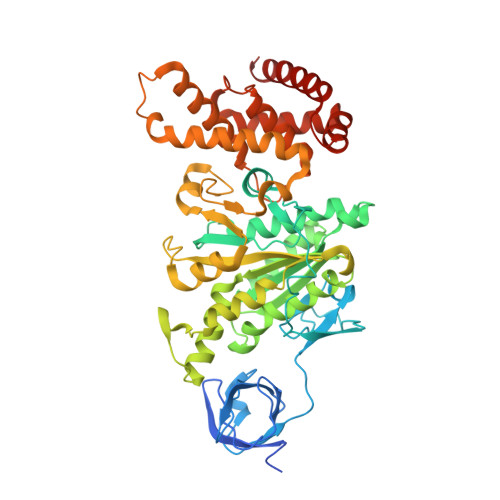
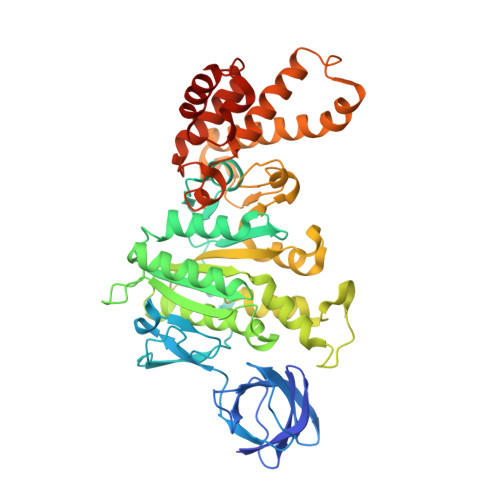
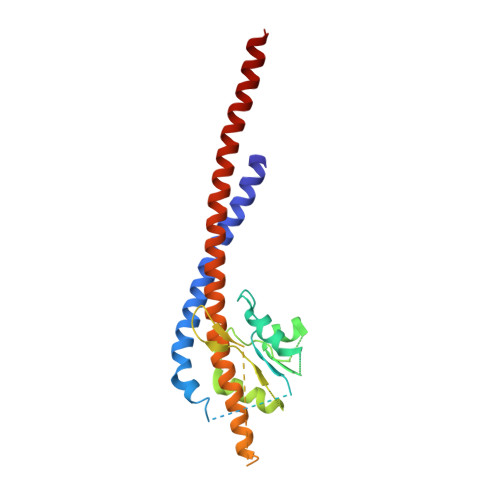
Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Gledhill, J.R., Montgomery, M.G., Leslie, A.G.W., Walker, J.E.(2007) Proc Natl Acad Sci U S A 104: 13632
- PubMed: 17698806
- DOI: https://doi.org/10.1073/pnas.0706290104
- Primary Citation of Related Structures:
2JIZ, 2JJ1, 2JJ2 - PubMed Abstract:
The structures of F(1)-ATPase from bovine heart mitochondria inhibited with the dietary phytopolyphenol, resveratrol, and with the related polyphenols quercetin and piceatannol have been determined at 2.3-, 2.4- and 2.7-A resolution, respectively. The inhibitors bind to a common site in the inside surface of an annulus made from loops in the three alpha- and three beta-subunits beneath the "crown" of beta-strands in their N-terminal domains. This region of F(1)-ATPase forms a bearing to allow the rotation of the tip of the gamma-subunit inside the annulus during catalysis. The binding site is a hydrophobic pocket between the C-terminal tip of the gamma-subunit and the beta(TP) subunit, and the inhibitors are bound via H-bonds mostly to their hydroxyl moieties mediated by bound water molecules and by hydrophobic interactions. There are no equivalent sites between the gamma-subunit and either the beta(DP) or the beta(E) subunit. The inhibitors probably prevent both the synthetic and hydrolytic activities of the enzyme by blocking both senses of rotation of the gamma-subunit. The beneficial effects of dietary resveratrol may derive in part by preventing mitochondrial ATP synthesis in tumor cells, thereby inducing apoptosis.
Organizational Affiliation:
Medical Research Council Dunn Human Nutrition Unit, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 0XY, United Kingdom.