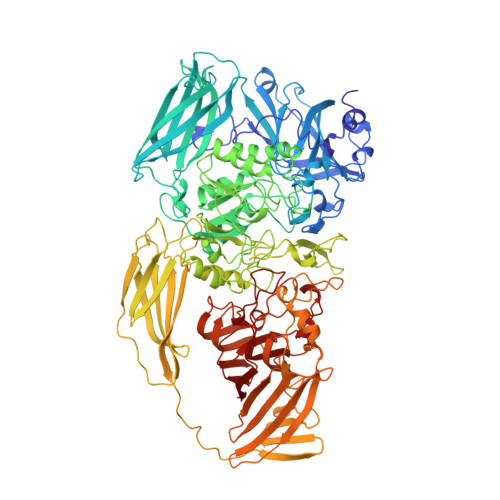
Three-dimensional structure of beta-galactosidase from E. coli.
Jacobson, R.H., Zhang, X.J., DuBose, R.F., Matthews, B.W.(1994) Nature 369: 761-766
- PubMed: 8008071
- DOI: https://doi.org/10.1038/369761a0
- Primary Citation of Related Structures:
4V40 - PubMed Abstract:
The beta-galactosidase from Escherichia coli was instrumental in the development of the operon model, and today is one of the most commonly used enzymes in molecular biology. Here we report the structure of this protein and show that it is a tetramer with 222-point symmetry. The 1,023-amino-acid polypeptide chain folds into five sequential domains, with an extended segment at the amino terminus. The participation of this amino-terminal segment in a subunit interface, coupled with the observation that each active site is made up of elements from two different subunits, provides a structural rationale for the phenomenon of alpha-complementation. The structure represents the longest polypeptide chain for which an atomic structure has been determined. Our results show that it is possible successfully to study non-viral protein crystals with unit cell dimensions in excess of 500 A and with relative molecular masses in the region of 2,000K per asymmetric unit. Non-crystallographic symmetry averaging proved to be a very powerful tool in the structure determination, as has been shown in other contexts.
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute, University of Oregon, Eugene 97403.