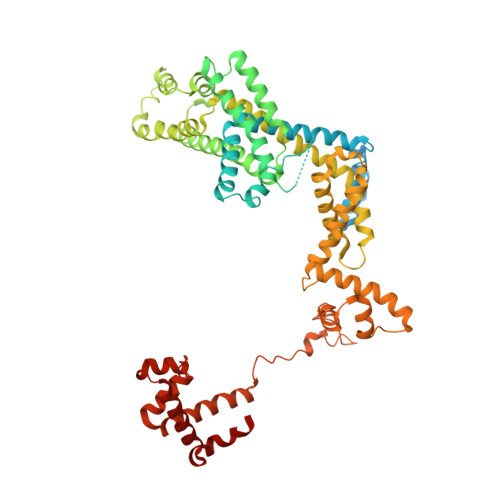
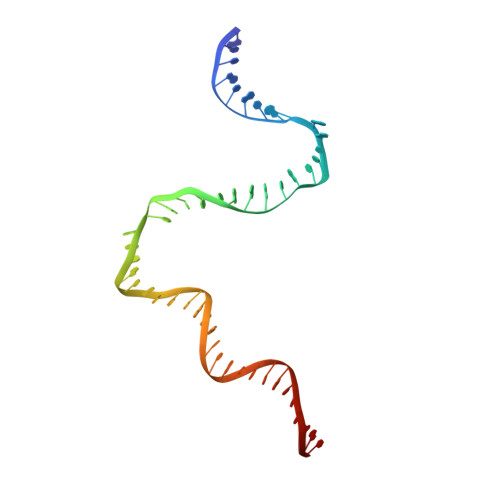
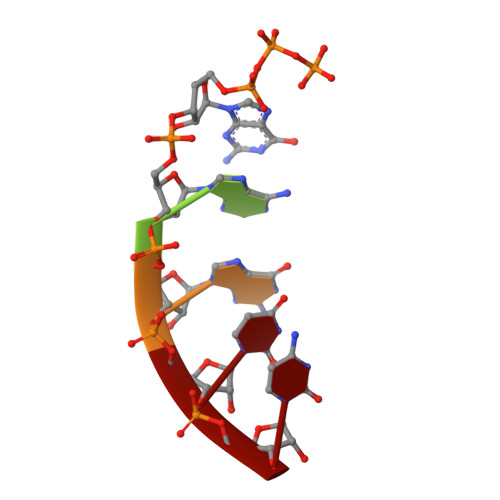
Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Zuo, Y., Steitz, T.A.(2015) Mol Cell 58: 534-540
- PubMed: 25866247
- DOI: https://doi.org/10.1016/j.molcel.2015.03.010
- Primary Citation of Related Structures:
4YLN, 4YLO, 4YLP - PubMed Abstract:
During transcription initiation, RNA polymerase binds to promoter DNA to form an initiation complex containing a DNA bubble and enters into abortive cycles of RNA synthesis before escaping the promoter to transit into the elongation phase for processive RNA synthesis. Here we present the crystal structures of E. coli transcription initiation complexes containing a complete transcription bubble and de novo synthesized RNA oligonucleotides at about 6-Å resolution. The structures show how RNA polymerase recognizes DNA promoters that contain spacers of different lengths and reveal a bridging interaction between the 5'-triphosphate of the nascent RNA and the σ factor that may function to stabilize the short RNA-DNA hybrids during the early stage of transcription initiation. The conformation of the RNA oligonucleotides and the paths of the DNA strands in the complete initiation complexes provide insights into the mechanism that controls both the abortive and productive RNA synthesis.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.