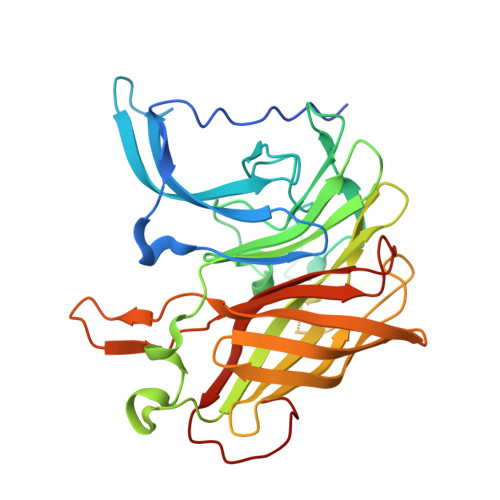
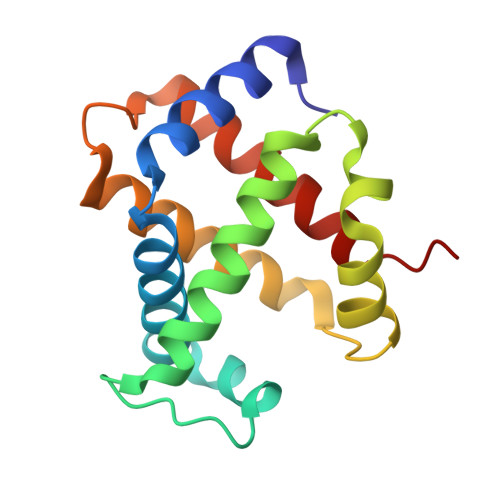
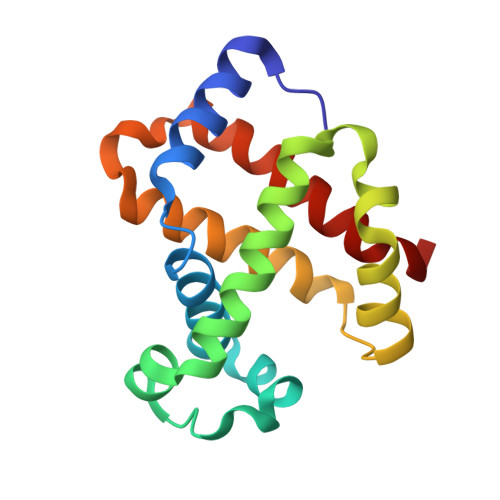
Structural analysis of haemoglobin binding by HpuA from the Neisseriaceae family.
Wong, C.T., Xu, Y., Gupta, A., Garnett, J.A., Matthews, S.J., Hare, S.A.(2015) Nat Commun 6: 10172-10172
- PubMed: 26671256
- DOI: https://doi.org/10.1038/ncomms10172
- Primary Citation of Related Structures:
5EC6, 5EE2, 5EE4 - PubMed Abstract:
The Neisseriaceae family of bacteria causes a range of diseases including meningitis, septicaemia, gonorrhoea and endocarditis, and extracts haem from haemoglobin as an important iron source within the iron-limited environment of its human host. Herein we report crystal structures of apo- and haemoglobin-bound HpuA, an essential component of this haem import system. The interface involves long loops on the bacterial receptor that present hydrophobic side chains for packing against the surface of haemoglobin. Interestingly, our structural and biochemical analyses of Kingella denitrificans and Neisseria gonorrhoeae HpuA mutants, although validating the interactions observed in the crystal structure, show how Neisseriaceae have the fascinating ability to diversify functional sequences and yet retain the haemoglobin binding function. Our results present the first description of HpuA's role in direct binding of haemoglobin.
Organizational Affiliation:
Department of Life Sciences, Imperial College London, South Kensington Campus, Exhibition Road, London SW7 2AZ, UK.