Design, Synthesis, and Cytotoxic Evaluation of Novel Tubulysin Analogues as ADC Payloads.
Leverett, C.A., Sukuru, S.C., Vetelino, B.C., Musto, S., Parris, K., Pandit, J., Loganzo, F., Varghese, A.H., Bai, G., Liu, B., Liu, D., Hudson, S., Doppalapudi, V.R., Stock, J., O'Donnell, C.J., Subramanyam, C.(2016) ACS Med Chem Lett 7: 999-1004
- PubMed: 27882198
- DOI: https://doi.org/10.1021/acsmedchemlett.6b00274
- Primary Citation of Related Structures:
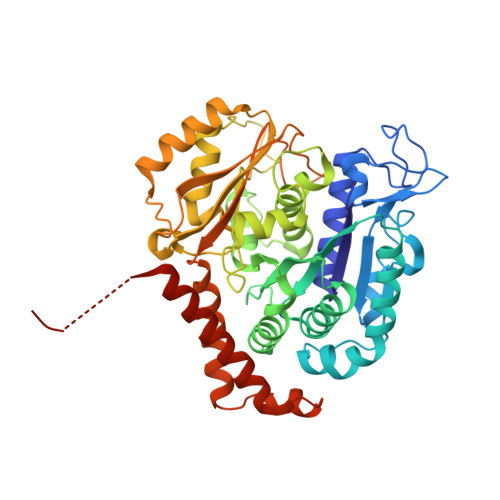
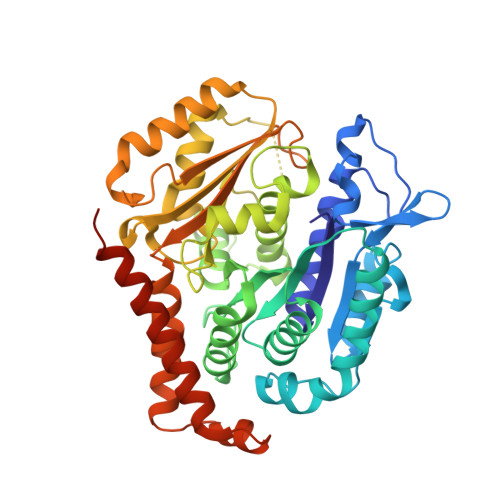
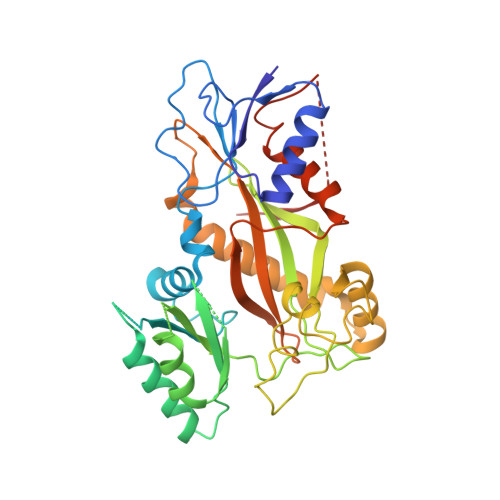
5KX5 - PubMed Abstract:
The tubulysin class of natural products has attracted much attention from the medicinal chemistry community due to its potent cytotoxicity against a wide range of human cancer cell lines, including significant activity in multidrug-resistant carcinoma models. As a result of their potency, the tubulysins have become an important tool for use in targeted therapy, being widely pursued as payloads in the development of novel small molecule drug conjugates (SMDCs) and antibody-drug conjugates (ADCs). A structure-based and parallel medicinal chemistry approach was applied to the synthesis of novel tubulysin analogues. These efforts led to the discovery of a number of novel and potent cytotoxic tubulysin analogues, providing a framework for our simultaneous report, which highlights the discovery of tubulysin-based ADCs, including use of site-specific conjugation to address in vivo stability of the C-11 acetate functionality.
Organizational Affiliation:
Medicinal Sciences, Pfizer Worldwide R&D , Groton, Connecticut 06340, United States.