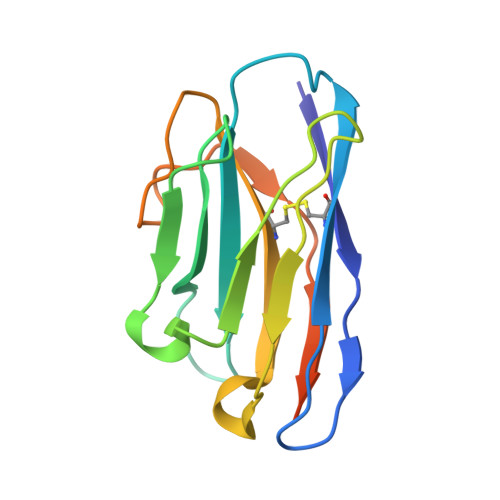
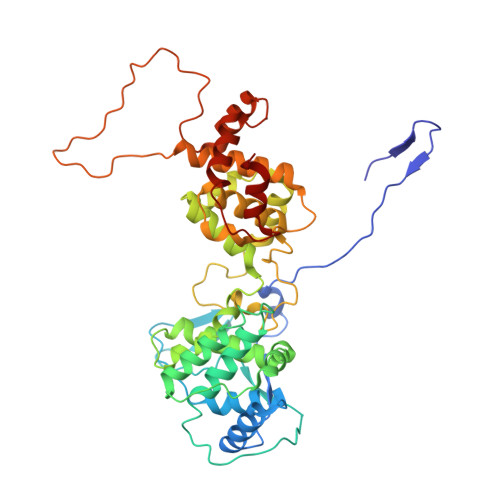
Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication.
Hanke, L., Schmidt, F.I., Knockenhauer, K.E., Morin, B., Whelan, S.P., Schwartz, T.U., Ploegh, H.L.(2017) EMBO Rep 18: 1027-1037
- PubMed: 28396572
- DOI: https://doi.org/10.15252/embr.201643764
- Primary Citation of Related Structures:
5UK4, 5UKB - PubMed Abstract:
The transcription and replication machinery of negative-stranded RNA viruses presents a possible target for interference in the viral life cycle. We demonstrate the validity of this concept through the use of cytosolically expressed single-domain antibody fragments (VHHs) that protect cells from a lytic infection with vesicular stomatitis virus (VSV) by targeting the viral nucleoprotein N. We define the binding sites for two such VHHs, 1004 and 1307, by X-ray crystallography to better understand their inhibitory properties. We found that VHH 1307 competes with the polymerase cofactor P for binding and thus inhibits replication and mRNA transcription, while binding of VHH 1004 likely only affects genome replication. The functional relevance of these epitopes is confirmed by the isolation of escape mutants able to replicate in the presence of the inhibitory VHHs. The escape mutations allow identification of the binding site of a third VHH that presumably competes with P for binding at another site than 1307. Collectively, these binding sites uncover different features on the N protein surface that may be suitable for antiviral intervention.
Organizational Affiliation:
Whitehead Institute for Biomedical Research, Cambridge, MA, USA.