Structural Basis for Selective Binding of Export Cargoes by Exportin-5
Yamazawa, R., Jiko, C., Choi, S., Park, I.Y., Nakagawa, A., Yamashita, E., Lee, S.J.(2018) Structure 26: 1393-1398.e2
- PubMed: 30100359
- DOI: https://doi.org/10.1016/j.str.2018.06.014
- Primary Citation of Related Structures:
5YU6, 5YU7 - PubMed Abstract:
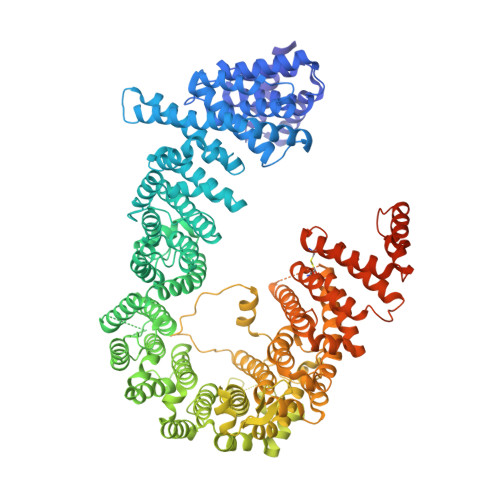
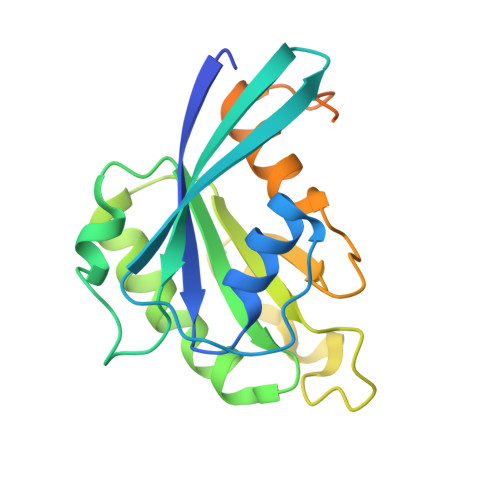
In the nucleus, RanGTP binding to importin dissociates the cargo. On the other hand, RanGTP enables exportin to bind export cargo and form the export complex by each exportin's own cargo selection mechanism. Here, we present two X-ray structures for Exportin-5 (Exp-5) alone and Exp-5:RanGTP intermediate complex. The structure of Exp-5 adopts a ring-shaped closed conformation by C-terminal anchor residues 1,167-1,179, interacting with N-terminal heat repeats 4-9. The closed form of Exp-5 is important for the stability of the cargo-free state. Interaction between Exp-5 and RanGTP induces elimination of intramolecular contacts of the C-terminal anchor. A large movement of N-terminal 1-9th heat repeats and C-terminal 19-20th heat repeats creates an open space for RanGTP accommodation. Exp-5 in Exp-5:RanGTP and Exp-5:RanGTP:pre-miRNA adopts the same conformation. RanGTP binding to Exp-5 creates a selective molecular cage area for accepting its cargoes, such as small double-stranded RNAs, without conformational change in Exp-5:RanGTP.
Organizational Affiliation:
Institute for Protein Research, Osaka University, 3-2 Yamada-oka, Suita, Osaka 565-0871, Japan.