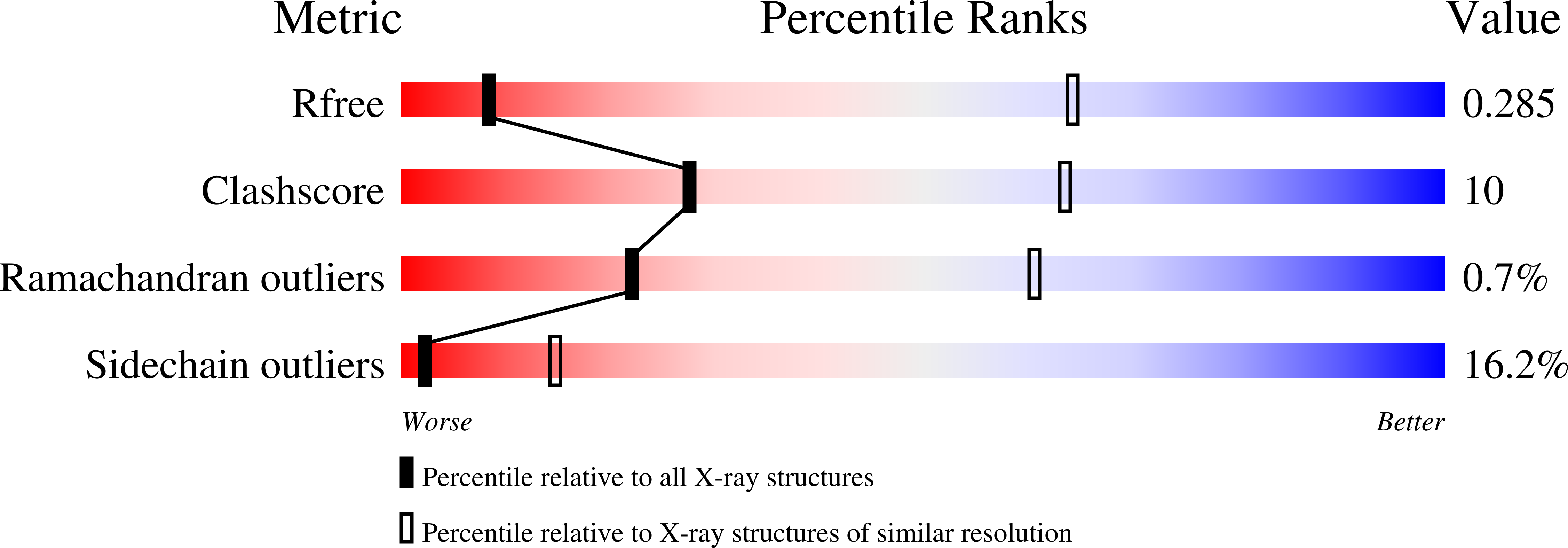
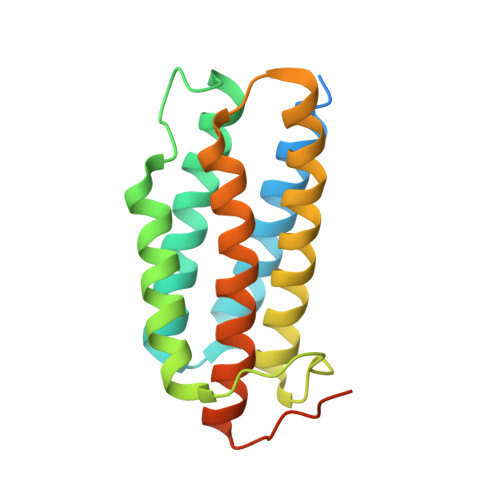
Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Cannon, K.A., Park, R.U., Boyken, S.E., Nattermann, U., Yi, S., Baker, D., King, N.P., Yeates, T.O.(2020) Protein Sci 29: 919-929
- PubMed: 31840320
- DOI: https://doi.org/10.1002/pro.3802
- Primary Citation of Related Structures:
5CY5, 5VL4 - PubMed Abstract:
In recent years, new protein engineering methods have produced more than a dozen symmetric, self-assembling protein cages whose structures have been validated to match their design models with near-atomic accuracy. However, many protein cage designs that are tested in the lab do not form the desired assembly, and improving the success rate of design has been a point of recent emphasis. Here we present two protein structures solved by X-ray crystallography of designed protein oligomers that form two-component cages with tetrahedral symmetry. To improve on the past tendency toward poorly soluble protein, we used a computational protocol that favors the formation of hydrogen-bonding networks over exclusively hydrophobic interactions to stabilize the designed protein-protein interfaces. Preliminary characterization showed highly soluble expression, and solution studies indicated successful cage formation by both designed proteins. For one of the designs, a crystal structure confirmed at high resolution that the intended tetrahedral cage was formed, though several flipped amino acid side chain rotamers resulted in an interface that deviates from the precise hydrogen-bonding pattern that was intended. A structure of the other designed cage showed that, under the conditions where crystals were obtained, a noncage structure was formed wherein a porous 3D protein network in space group I2 1 3 is generated by an off-target twofold homomeric interface. These results illustrate some of the ongoing challenges of developing computational methods for polar interface design, and add two potentially valuable new entries to the growing list of engineered protein materials for downstream applications.
Organizational Affiliation:
UCLA-DOE Institute for Genomics and Proteomics, Los Angeles, California.