Cobalt complex structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subsp. spizizenii.
Nam, M.S., Song, W.S., Park, S.C., Yoon, S.I.(2019) Korean J Microbiol 55: 123-130
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2019) Korean J Microbiol 55: 123-130
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sirohydrochlorin ferrochelatase | 267 | Bacillus spizizenii | Mutation(s): 0 Gene Names: sirB EC: 4.99.1.3 (PDB Primary Data), 4.99.1.4 (PDB Primary Data) | 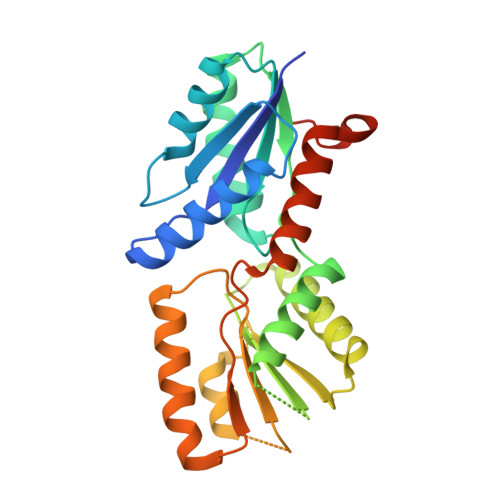 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CO Query on CO | B [auth A], C [auth A] | COBALT (II) ION Co XLJKHNWPARRRJB-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.669 | α = 90 |
| b = 75.669 | β = 90 |
| c = 82.419 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |