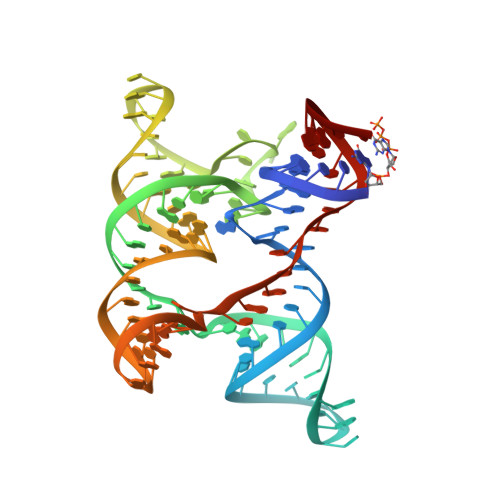
Local-to-global signal transduction at the core of a Mn2+sensing riboswitch.
Suddala, K.C., Price, I.R., Dandpat, S.S., Janecek, M., Kuhrova, P., Sponer, J., Banas, P., Ke, A., Walter, N.G.(2019) Nat Commun 10: 4304-4304
- PubMed: 31541094
- DOI: https://doi.org/10.1038/s41467-019-12230-5
- Primary Citation of Related Structures:
6N2V - PubMed Abstract:
The widespread Mn 2+ -sensing yybP-ykoY riboswitch controls the expression of bacterial Mn 2+ homeostasis genes. Here, we first determine the crystal structure of the ligand-bound yybP-ykoY riboswitch aptamer from Xanthomonas oryzae at 2.96 Å resolution, revealing two conformations with docked four-way junction (4WJ) and incompletely coordinated metal ions. In >100 µs of MD simulations, we observe that loss of divalents from the core triggers local structural perturbations in the adjacent docking interface, laying the foundation for signal transduction to the regulatory switch helix. Using single-molecule FRET, we unveil a previously unobserved extended 4WJ conformation that samples transient docked states in the presence of Mg 2+ . Only upon adding sub-millimolar Mn 2+ , however, can the 4WJ dock stably, a feature lost upon mutation of an adenosine contacting Mn 2+ in the core. These observations illuminate how subtly differing ligand preferences of competing metal ions become amplified by the coupling of local with global RNA dynamics.
Organizational Affiliation:
Single Molecule Analysis Group and Center for RNA Biomedicine, Department of Chemistry, University of Michigan, Ann Arbor, MI, 48109, USA.