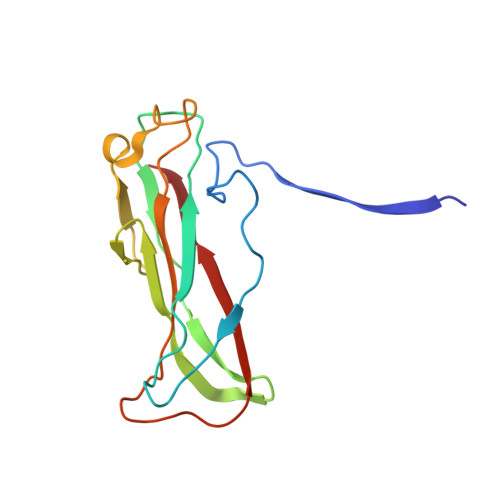
Cryo-EM structure of the CFA/I pilus rod.
Zheng, W., Andersson, M., Mortezaei, N., Bullitt, E., Egelman, E.(2019) IUCrJ 6: 815-821
- PubMed: 31576215
- DOI: https://doi.org/10.1107/S2052252519007966
- Primary Citation of Related Structures:
6NRV - PubMed Abstract:
Enterotoxigenic Escherichia coli (ETEC) are common agents of diarrhea for travelers and a major cause of mortality in children in developing countries. To attach to intestinal cells ETEC express colonization factors, among them CFA/I, which are the most prevalent factors and are the archetypical representative of class 5 pili. The helical quaternary structure of CFA/I can be unwound under tensile force and it has been shown that this mechanical property helps bacteria to withstand shear forces from fluid motion. We report in this work the CFA/I pilus structure at 4.3 Å resolution from electron cryomicroscopy (cryo-EM) data, and report details of the donor strand complementation. The CfaB pilins modeled into the cryo-EM map allow us to identify the buried surface area between subunits, and these regions are correlated to quaternary structural stability in class 5 and chaperone-usher pili. In addition, from the model built using the EM structure we also predicted that residue 13 (proline) of the N-terminal β-strand could have a major impact on the filament's structural stability. Therefore, we used optical tweezers to measure and compare the stability of the quaternary structure of wild type CFA/I and a point-mutated CFA/I with a propensity for unwinding. We found that pili with this mutated CFA/I require a lower force to unwind, supporting our hypothesis that Pro13 is important for structural stability. The high-resolution CFA/I pilus structure presented in this work and the analysis of structural stability will be useful for the development of novel antimicrobial drugs that target adhesion pili needed for initial attachment and sustained adhesion of ETEC.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA, USA.