Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Huhmann, S., Nyakatura, E.K., Rohrhofer, A., Moschner, J., Schmidt, B., Eichler, J., Roth, C., Koksch, B.(2021) Chembiochem 22: 3443-3451
- PubMed: 34605595
- DOI: https://doi.org/10.1002/cbic.202100417
- Primary Citation of Related Structures:
6TVQ, 6TVU, 6TVW - PubMed Abstract:
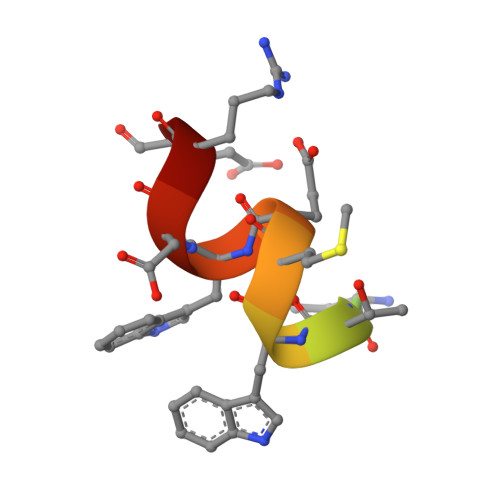
With the emergence of novel viruses, the development of new antivirals is more urgent than ever. A key step in human immunodeficiency virus type 1 (HIV-1) infection is six-helix bundle formation within the envelope protein subunit gp41. Selective disruption of bundle formation by peptides has been shown to be effective; however, these drugs, exemplified by T20, are prone to rapid clearance from the patient. The incorporation of non-natural amino acids is known to improve these pharmacokinetic properties. Here, we evaluate a peptide inhibitor in which a critical Ile residue is replaced by fluorinated analogues. We characterized the influence of the fluorinated analogues on the biophysical properties of the peptide. Furthermore, we show that the fluorinated peptides can block HIV-1 infection of target cells at nanomolar levels. These findings demonstrate that fluorinated amino acids are appropriate tools for the development of novel peptide therapeutics.
Organizational Affiliation:
Freie Universität Berlin, Department of Biology, Chemistry and Pharmacy, Institute of Chemistry and Biochemistry, Arnimallee 20, 14195, Berlin, Germany.