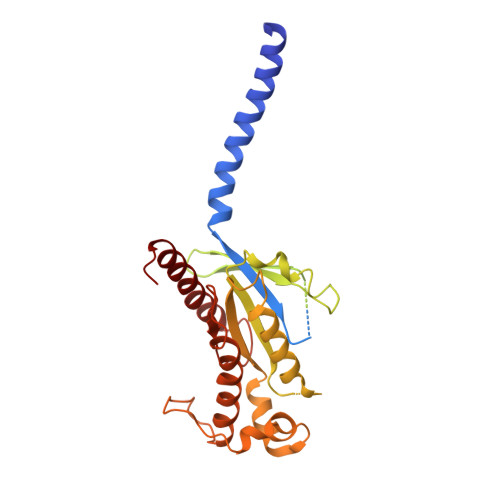
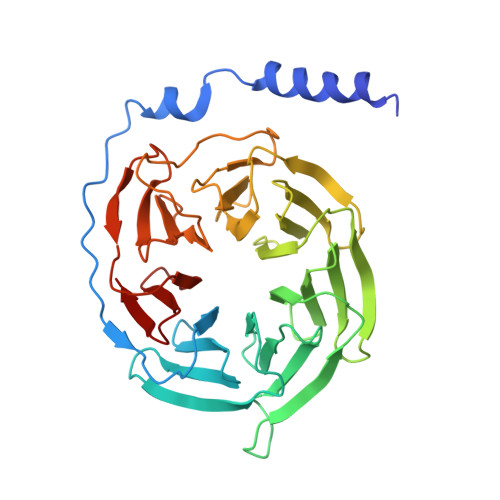
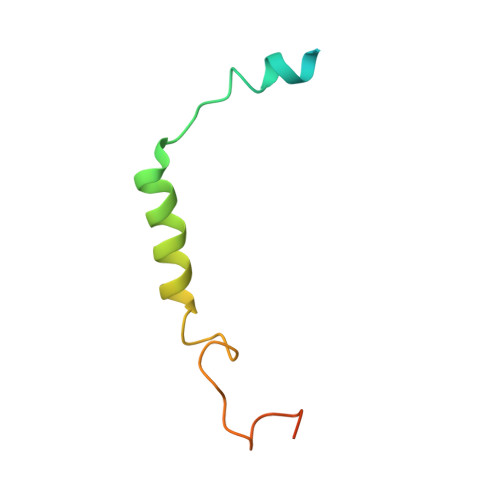
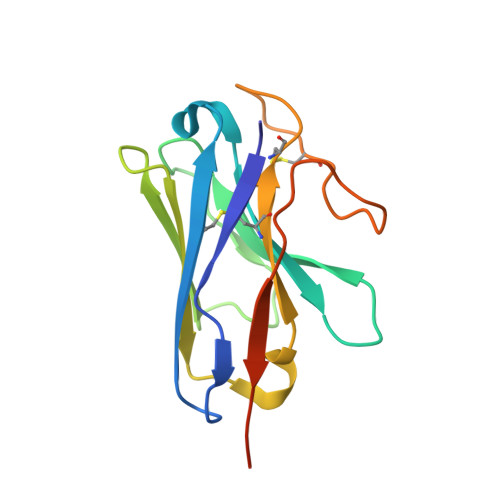
Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
Chang, R., Zhang, X., Qiao, A., Dai, A., Belousoff, M.J., Tan, Q., Shao, L., Zhong, L., Lin, G., Liang, Y.L., Ma, L., Han, S., Yang, D., Danev, R., Wang, M.W., Wootten, D., Wu, B., Sexton, P.M.(2020) J Biol Chem 295: 9313-9325
- PubMed: 32371397
- DOI: https://doi.org/10.1074/jbc.RA120.013793
- Primary Citation of Related Structures:
6WHC - PubMed Abstract:
Unimolecular dual agonists of the glucagon (GCG) receptor (GCGR) and glucagon-like peptide-1 receptor (GLP-1R) are a new class of drugs that are potentially superior to GLP-1R-specific agonists for the management of metabolic disease. The dual-agonist, peptide 15 (P15), is a glutamic acid 16 analog of GCG with GLP-1 peptide substitutions between amino acids 17 and 24 that has potency equivalent to those of the cognate peptide agonists at the GCGR and GLP-1R. Here, we have used cryo-EM to solve the structure of an active P15-GCGR-G s complex and compared this structure to our recently published structure of the GCGR-G s complex bound to GCG. This comparison revealed that P15 has a reduced interaction with the first extracellular loop (ECL1) and the top of transmembrane segment 1 (TM1) such that there is increased mobility of the GCGR extracellular domain and at the C terminus of the peptide compared with the GCG-bound receptor. We also observed a distinct conformation of ECL3 and could infer increased mobility of the far N-terminal His-1 residue in the P15-bound structure. These regions of conformational variance in the two peptide-bound GCGR structures were also regions that were distinct between GCGR structures and previously published peptide-bound structures of the GLP-1R, suggesting that greater conformational dynamics may contribute to the increased efficacy of P15 in activation of the GLP-1R compared with GCG. The variable domains in this receptor have previously been implicated in biased agonism at the GLP-1R and could result in altered signaling of P15 at the GCGR compared with GCG.
Organizational Affiliation:
School of Pharmacy, Shanghai Medical College, Fudan University, Shanghai, China.