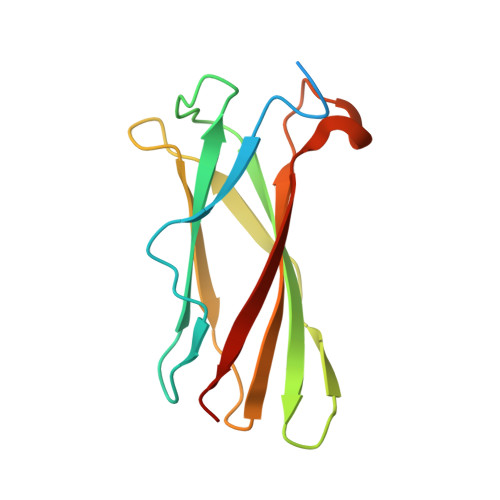
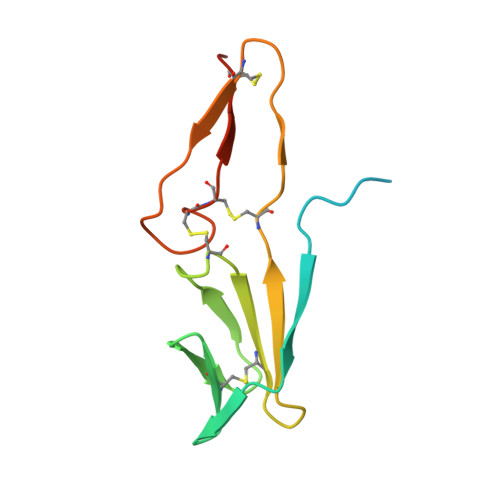
An inhibitor of complement C5 provides structural insights into activation.
Reichhardt, M.P., Johnson, S., Tang, T., Morgan, T., Tebeka, N., Popitsch, N., Deme, J.C., Jore, M.M., Lea, S.M.(2020) Proc Natl Acad Sci U S A 117: 362-370
- PubMed: 31871188
- DOI: https://doi.org/10.1073/pnas.1909973116
- Primary Citation of Related Structures:
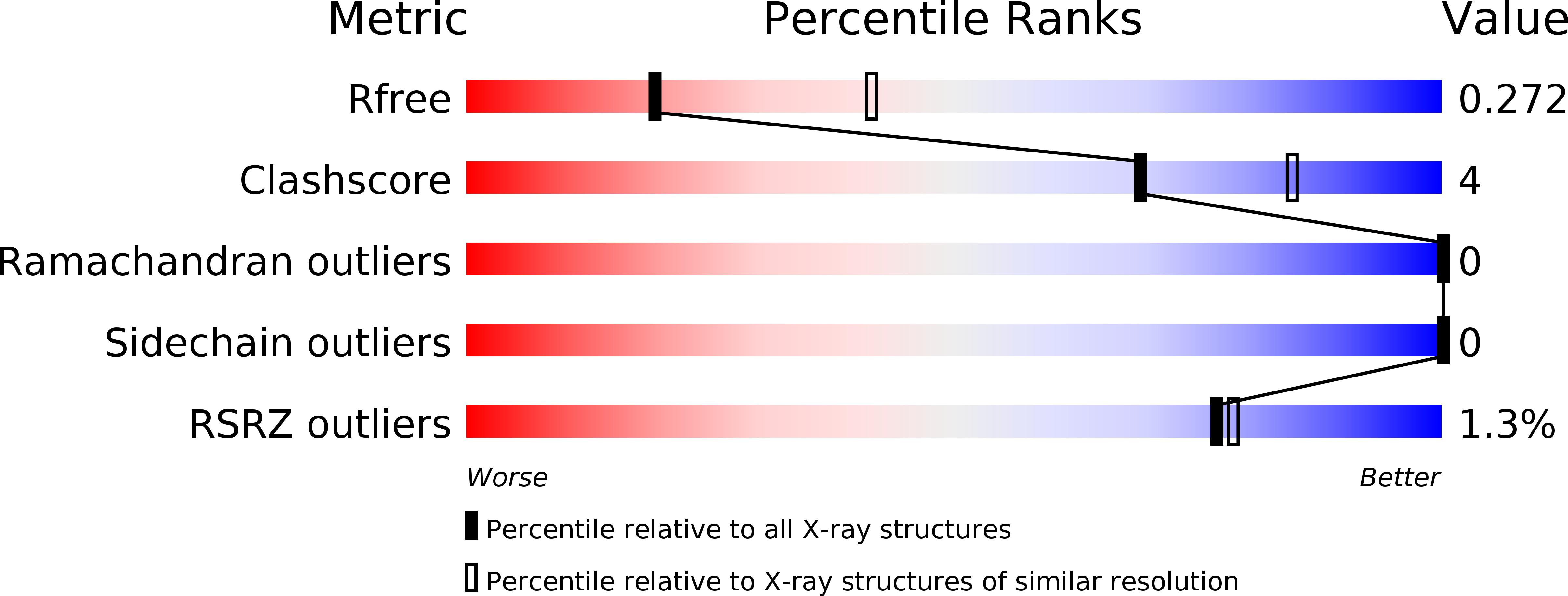
6RPT, 6RQJ - PubMed Abstract:
The complement system is a crucial part of innate immune defenses against invading pathogens. The blood-meal of the tick Rhipicephalus pulchellus lasts for days, and the tick must therefore rely on inhibitors to counter complement activation. We have identified a class of inhibitors from tick saliva, the CirpT family, and generated detailed structural data revealing their mechanism of action. We show direct binding of a CirpT to complement C5 and have determined the structure of the C5-CirpT complex by cryoelectron microscopy. This reveals an interaction with the peripheral macro globulin domain 4 (C5_MG4) of C5. To achieve higher resolution detail, the structure of the C5_MG4-CirpT complex was solved by X-ray crystallography (at 2.7 Å). We thus present the fold of the CirpT protein family, and provide detailed mechanistic insights into its inhibitory function. Analysis of the binding interface reveals a mechanism of C5 inhibition, and provides information to expand our biological understanding of the activation of C5, and thus the terminal complement pathway.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, OX1 3RE Oxford, United Kingdom.