Molecular insights into the human CLC-7/Ostm1 transporter.
Zhang, S., Liu, Y., Zhang, B., Zhou, J., Li, T., Liu, Z., Li, Y., Yang, M.(2020) Sci Adv 6: eabb4747-eabb4747
- PubMed: 32851177
- DOI: https://doi.org/10.1126/sciadv.abb4747
- Primary Citation of Related Structures:
7BXU - PubMed Abstract:
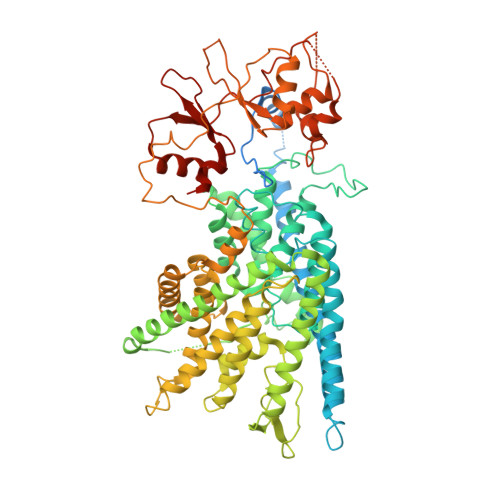
CLC family proteins translocate chloride ions across cell membranes to maintain the membrane potential, regulate the transepithelial Cl - transport, and control the intravesicular pH among different organelles. CLC-7/Ostm1 is an electrogenic Cl - /H + antiporter that mainly resides in lysosomes and osteoclast ruffled membranes. Mutations in human CLC-7/Ostm1 lead to lysosomal storage disorders and severe osteopetrosis. Here, we present the cryo-electron microscopy (cryo-EM) structure of the human CLC-7/Ostm1 complex and reveal that the highly glycosylated Ostm1 functions like a lid positioned above CLC-7 and interacts extensively with CLC-7 within the membrane. Our complex structure reveals a functionally crucial domain interface between the amino terminus, TMD, and CBS domains of CLC-7. Structural analyses and electrophysiology studies suggest that the domain interaction interfaces affect the slow gating kinetics of CLC-7/Ostm1. Thus, our study deepens understanding of CLC-7/Ostm1 transporter and provides insights into the molecular basis of the disease-related mutations.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.