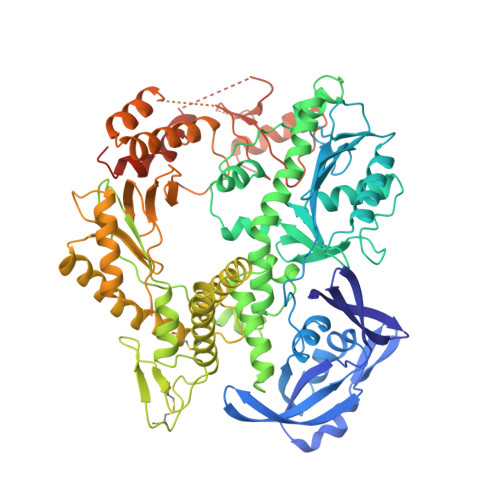
Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7.
Rodriguez, A.C., Park, H.W., Mao, C., Beese, L.S.(2000) J Mol Biol 299: 447-462
- PubMed: 10860752
- DOI: https://doi.org/10.1006/jmbi.2000.3728
- Primary Citation of Related Structures:
1QHT - PubMed Abstract:
The 2.25 A resolution crystal structure of a pol alpha family (family B) DNA polymerase from the hyperthermophilic marine archaeon Thermococcus sp. 9 degrees N-7 (9 degrees N-7 pol) provides new insight into the mechanism of pol alpha family polymerases that include essentially all of the eukaryotic replicative and viral DNA polymerases. The structure is folded into NH(2)- terminal, editing 3'-5' exonuclease, and polymerase domains that are topologically similar to the two other known pol alpha family structures (bacteriophage RB69 and the recently determined Thermococcus gorgonarius), but differ in their relative orientation and conformation. The 9 degrees N-7 polymerase domain structure is reminiscent of the "closed" conformation characteristic of ternary complexes of the pol I polymerase family obtained in the presence of their dNTP and DNA substrates. In the apo-9 degrees N-7 structure, this conformation appears to be stabilized by an ion pair. Thus far, the other apo-pol alpha structures that have been determined adopt open conformations. These results therefore suggest that the pol alpha polymerases undergo a series of conformational transitions during the catalytic cycle similar to those proposed for the pol I family. Furthermore, comparison of the orientations of the fingers and exonuclease (sub)domains relative to the palm subdomain that contains the pol active site suggests that the exonuclease domain and the fingers subdomain of the polymerase can move as a unit and may do so as part of the catalytic cycle. This provides a possible structural explanation for the interdependence of polymerization and editing exonuclease activities unique to pol alpha family polymerases. We suggest that the NH(2)-terminal domain of 9 degrees N-7 pol may be structurally related to an RNA-binding motif, which appears to be conserved among archaeal polymerases. The presence of such a putative RNA- binding domain suggests a mechanism for the observed autoregulation of bacteriophage T4 DNA polymerase synthesis by binding to its own mRNA. Furthermore, conservation of this domain could indicate that such regulation of pol expression may be a characteristic of archaea. Comparion of the 9 degrees N-7 pol structure to its mesostable homolog from bacteriophage RB69 suggests that thermostability is achieved by shortening loops, forming two disulfide bridges, and increasing electrostatic interactions at subdomain interfaces.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Durham, NC, 27710, USA.