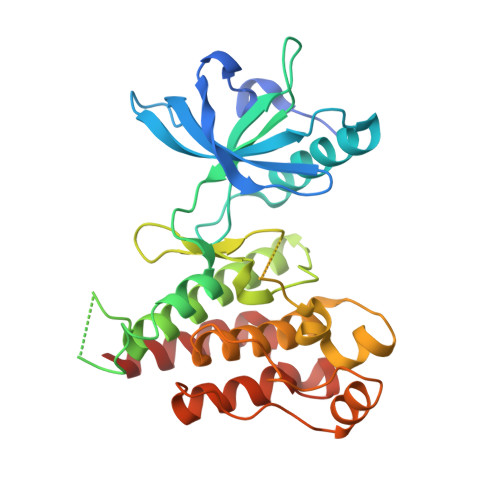
Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis.
McTigue, M.A., Wickersham, J.A., Pinko, C., Showalter, R.E., Parast, C.V., Tempczyk-Russell, A., Gehring, M.R., Mroczkowski, B., Kan, C.C., Villafranca, J.E., Appelt, K.(1999) Structure 7: 319-330
- PubMed: 10368301
- DOI: https://doi.org/10.1016/s0969-2126(99)80042-2
- Primary Citation of Related Structures:
1VR2 - PubMed Abstract:
Angiogenesis is involved in tumor growth, macular degeneration, retinopathy and other diseases. Vascular endothelial growth factor (VEGF) stimulates angiogenesis by binding to specific receptors (VEGFRs) on the surface of vascular endothelial cells. VEGFRs are receptor tyrosine kinases that, like the platelet-derived growth factor receptors (PDGFRs), contain a large insert within the kinase domain. We report here the generation, kinetic characterization, and 2.4 A crystal structure of the catalytic kinase domain of VEGF receptor 2 (VEGFR2). This protein construct, which lacks 50 central residues of the 68-residue kinase insert domain (KID), has comparable kinase activity to constructs containing the entire KID. The crystal structure, determined in an unliganded phosphorylated state, reveals an overall fold and catalytic residue positions similar to those observed in other tyrosine-kinase structures. The kinase activation loop, autophosphorylated on Y1059 prior to crystallization, is mostly disordered; however, a portion of it occupies a position inhibitory to substrate binding. The ends of the KID form a beta-like structure, not observed in other known tyrosine kinase structures, that packs near to the kinase C terminus. The majority of the VEGFR2 KID residues are not necessary for kinase activity. The unique structure observed for the ends of the KID may also occur in other PDGFR family members and may serve to properly orient the KID for signal transduction. This VEGFR2 kinase structure provides a target for design of selective anti-angiogenic therapeutic agents.
Organizational Affiliation:
Agouron Pharmaceuticals, 3565 General Atomics Court, San Diego, CA 92121, USA. mctigue@agouron.com