Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
Lillington, J.E.D., Lovett, J.E., Johnson, S., Roversi, P., Timmel, C.R., Lea, S.M.(2011) J Mol Biol 405: 427
- PubMed: 21075116
- DOI: https://doi.org/10.1016/j.jmb.2010.10.053
- Primary Citation of Related Structures:
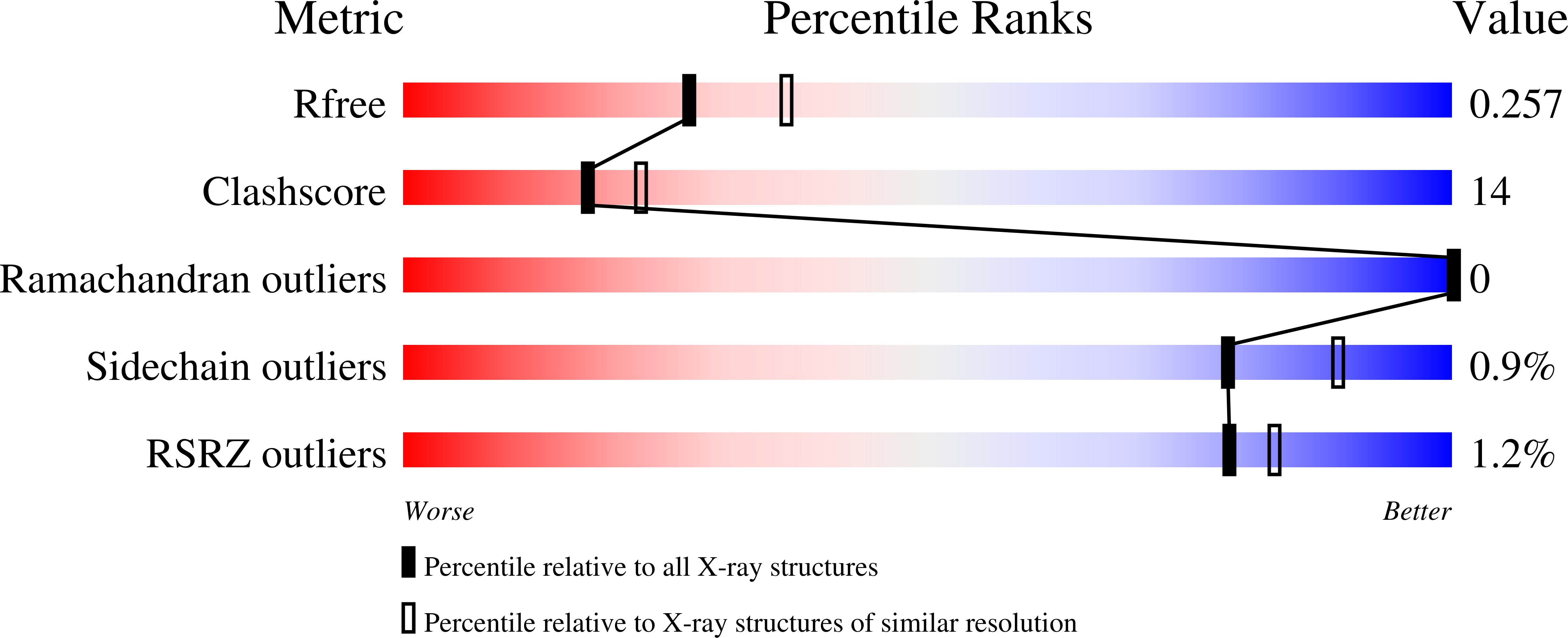
2XGA - PubMed Abstract:
Shigella flexneri Spa15 is a chaperone of the type 3 secretion system, which binds a number of effectors to ensure their stabilization prior to secretion. One of these effectors is IpgB1, a mimic of the human Ras-like Rho guanosine triphosphatase RhoG. In this study, Spa15 alone and in complex with IpgB1 has been studied by double electron electron resonance, an experiment that gives distance information showing the spacial separation of attached spin labels. This distance is explained by determining the crystal structure of the spin-labeled Spa15 where labels are seen to be buried in hydrophobic pockets. The double electron electron resonance experiment on the Spa15 complex with IpgB1 shows that IpgB1 does not bind Spa15 in the same way as is seen in the homologous Salmonella sp. chaperone:effector complex InvB:SipA.
Organizational Affiliation:
Inorganic Chemistry Laboratory, University of Oxford, OX1 3QR, UK.