The crystal structure of A SAM-Dependent enzyme from Aspergillus flavus
Zhou, Y.Z., Peng, T., Liao, L.J., Zhao, Y.C., Liu, X.K., Guo, Y., Zeng, Z.X.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| methyltransferase lepI | 386 | Aspergillus flavus NRRL3357 | Mutation(s): 0 Gene Names: lepI, AFLA_066940 EC: 2.1.1 | 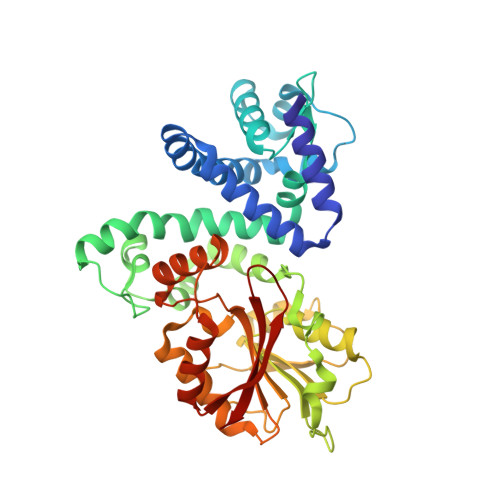 | |
UniProt | |||||
Find proteins for B8NJH3 (Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167)) Explore B8NJH3 Go to UniProtKB: B8NJH3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B8NJH3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAH Query on SAH | C [auth A], D [auth B] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 160.556 | α = 90 |
| b = 62.225 | β = 113.06 |
| c = 113.354 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHENIX | phasing |