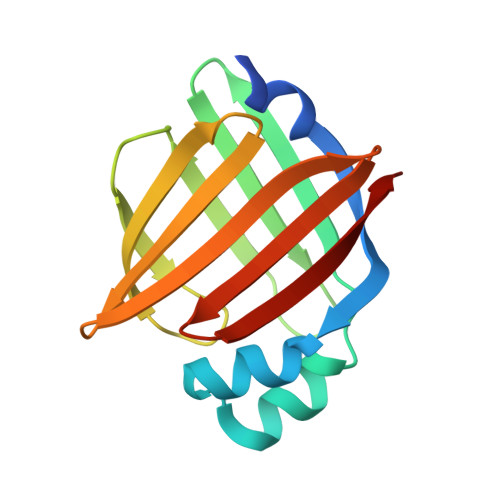
Structural Insights into Mouse H-FABP.
Wang, L., Zhang, H., Lv, P., Li, Y., Teng, M., Liu, Y., Wu, D.(2022) Life (Basel) 12
- PubMed: 36143481
- DOI: https://doi.org/10.3390/life12091445
- Primary Citation of Related Structures:
7YF1 - PubMed Abstract:
Intracellular fatty acid-binding proteins are evolutionarily highly conserved proteins. The major functions and responsibilities of this family are the regulation of FA uptake and intracellular transport. The structure of the H-FABP ortholog from mouse ( Mus musculus ) had not been revealed at the time this study was completed. Thus, further exploration of the structural properties of mouse H-FABP is expected to extend our knowledge of the model animal's molecular mechanism of H-FABP function. Here, we report the high-resolution crystal structure and the NMR characterization of mouse H-FABP. Our work discloses the unique structural features of mouse H-FABP, offering a structural basis for the further development of small-molecule inhibitors for H-FABP.
Organizational Affiliation:
School of Life Science, University of Science and Technology of China, Hefei 230027, China.