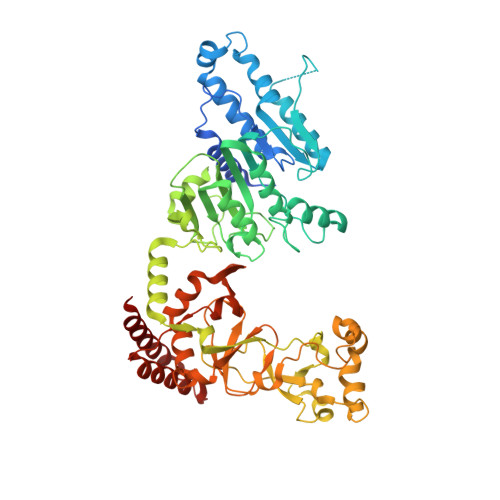
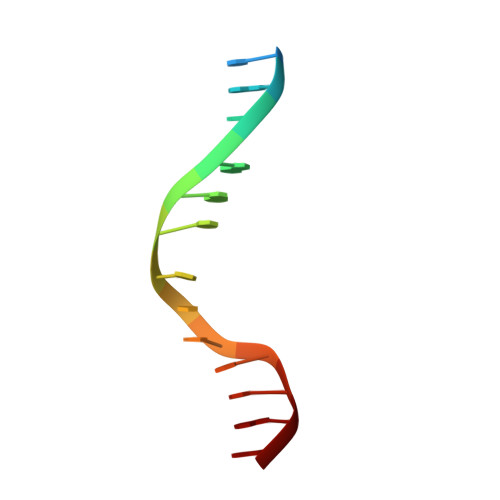
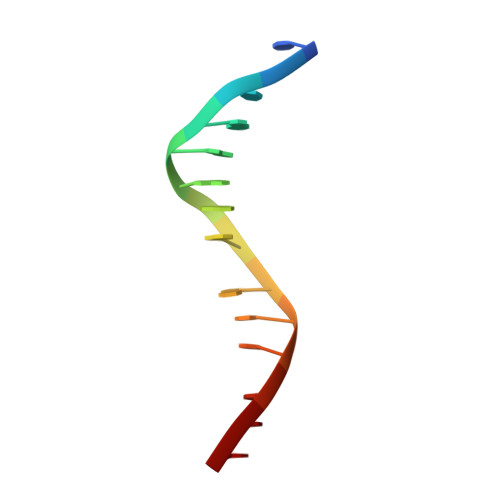
Quinoline-based compounds can inhibit diverse enzymes that act on DNA
Zhou, J., Chen, Q., Ren, R., Yang, J., Chang, C., Liu, B., Horton, J.R., Li, C., Maksoud, L., Yang, Y., Rotili, D., Jain, A.K., Zhang, X., Blumenthal, R.M., Chen, T., Gao, Y., Valente, S., Mai, A., Cheng, X.(2024) Cell Chem Biol