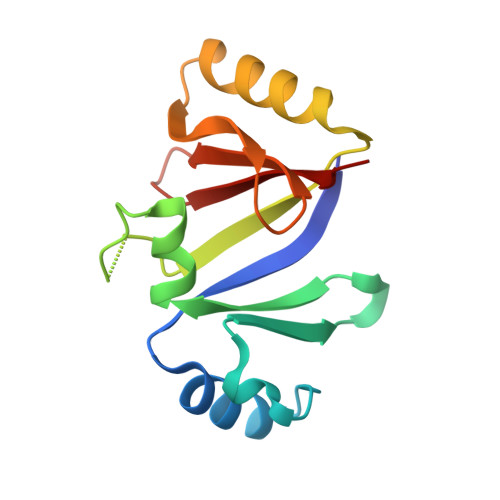
Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Martin, T.W., Dauter, Z., Devedjiev, Y., Sheffield, P., Jelen, F., He, M., Sherman, D.H., Otlewski, J., Derewenda, Z.S., Derewenda, U.(2002) Structure 10: 933-942
- PubMed: 12121648
- DOI: https://doi.org/10.1016/s0969-2126(02)00778-5
- Primary Citation of Related Structures:
1KLL, 1KMZ - PubMed Abstract:
Mitomycin C (MC) is a potent anticancer agent. Streptomyces lavendulae, which produces MC, protects itself from the lethal effects of the drug by expressing several resistance proteins. One of them (MRD) binds MC and functions as a drug exporter. We report the crystal structure of MRD and its complex with an MC metabolite, 1,2-cis-1-hydroxy-2,7-diaminomitosene, at 1.5 A resolution. The drug is sandwiched by pi-stacking interactions of His-38 and Trp-108. MRD is a dimer. The betaalphabetabetabeta fold of the MRD molecule is reminiscent of methylmalonyl-CoA epimerase, bleomycin resistance proteins, glyoxalase I, and extradiol dioxygenases. The location of the binding site is identical to the ones in evolutionarily related enzymes, suggesting that the protein may have been recruited from a different metabolic pathway.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Health Sciences System, Charlottesville 22908, USA.