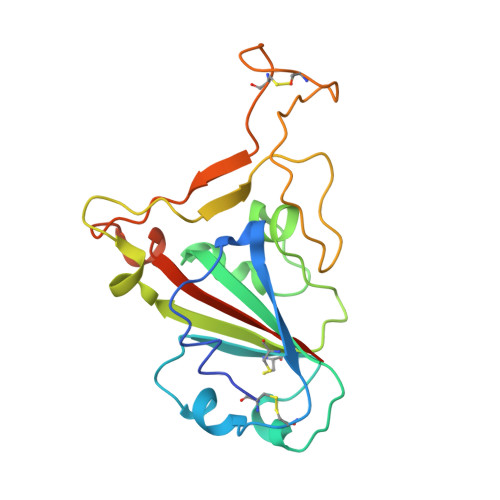
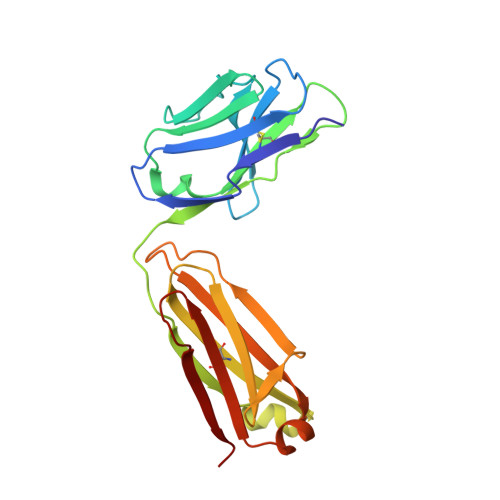
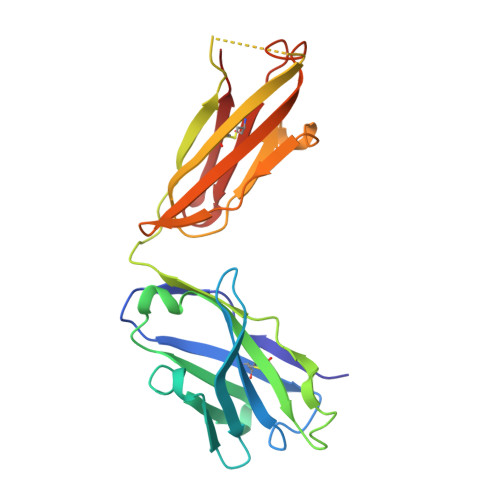
Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain.
Pak, J.E., Sharon, C., Satkunarajah, M., Auperin, T.C., Cameron, C.M., Kelvin, D.J., Seetharaman, J., Cochrane, A., Plummer, F.A., Berry, J.D., Rini, J.M.(2009) J Mol Biol 388: 815-823
- PubMed: 19324051
- DOI: https://doi.org/10.1016/j.jmb.2009.03.042
- Primary Citation of Related Structures:
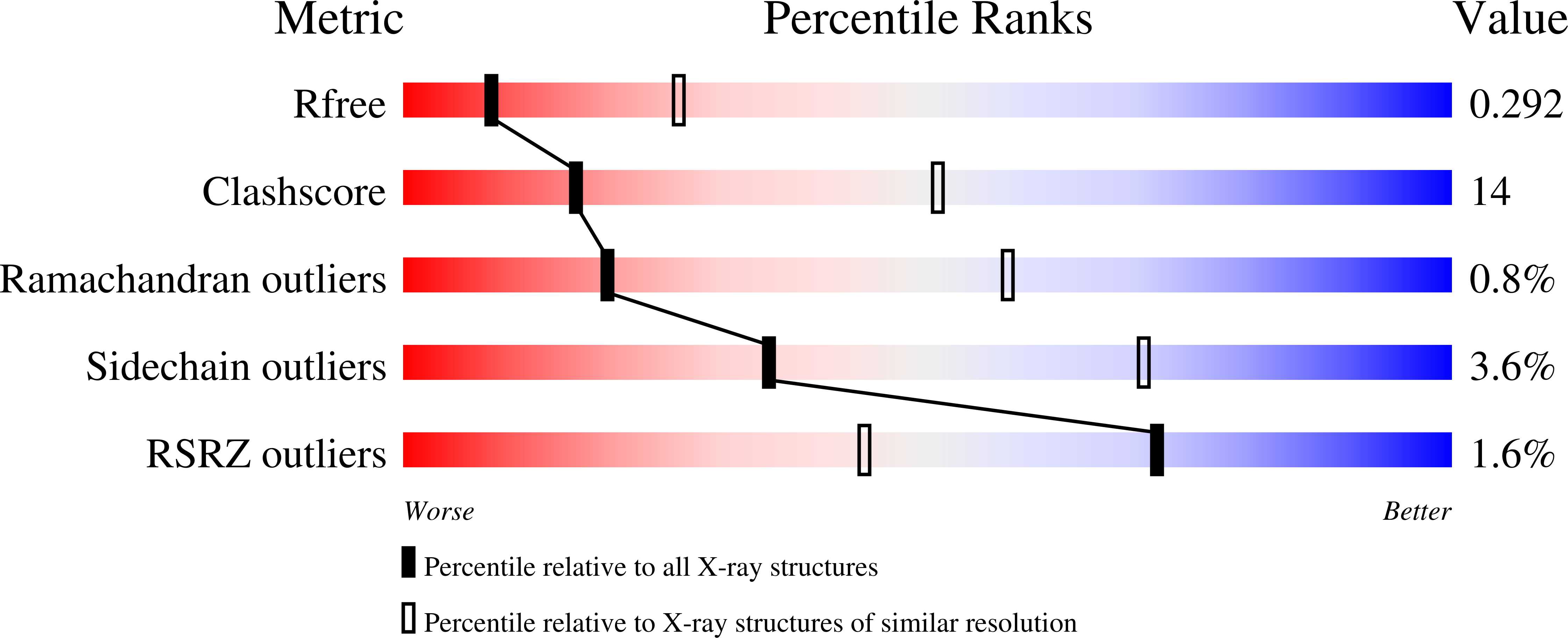
3BGF - PubMed Abstract:
The spike (S) protein of the severe acute respiratory syndrome coronavirus (SARS-CoV) is responsible for host cell attachment and fusion of the viral and host cell membranes. Within S the receptor binding domain (RBD) mediates the interaction with angiotensin-converting enzyme 2 (ACE2), the SARS-CoV host cell receptor. Both S and the RBD are highly immunogenic and both have been found to elicit neutralizing antibodies. Reported here is the X-ray crystal structure of the RBD in complex with the Fab of a neutralizing mouse monoclonal antibody, F26G19, elicited by immunization with chemically inactivated SARS-CoV. The RBD-F26G19 Fab complex represents the first example of the structural characterization of an antibody elicited by an immune response to SARS-CoV or any fragment of it. The structure reveals that the RBD surface recognized by F26G19 overlaps significantly with the surface recognized by ACE2 and, as such, suggests that F26G19 likely neutralizes SARS-CoV by blocking the virus-host cell interaction.
Organizational Affiliation:
Department of Molecular Genetics, University of Toronto, Toronto, Ontario, Canada.