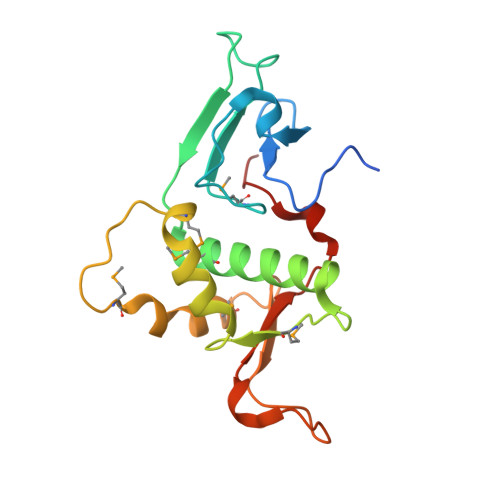
Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding.
Chen, Y., Wan, B., Wang, K.C., Cao, F., Yang, Y., Protacio, A., Dou, Y., Chang, H.Y., Lei, M.(2011) EMBO Rep 12: 797-803
- PubMed: 21660059
- DOI: https://doi.org/10.1038/embor.2011.101
- Primary Citation of Related Structures:
3RSN - PubMed Abstract:
Ash2L is a core component of the MLL family histone methyltransferases and has an important role in regulating the methylation of histone H3 on lysine 4. Here, we report the crystal structure of the N-terminal domain of Ash2L and reveal a new function of Ash2L. The structure shows that Ash2L contains an atypical PHD finger that does not have histone tail-binding activity. Unexpectedly, the structure shows a previously unrecognized winged-helix motif that directly binds to DNA. The DNA-binding-deficient mutants of Ash2L reduced Ash2L localization to the HOX locus. Strikingly, a single mutation in Ash2L(WH) (K131A) breaks the chromatin domain boundary, suggesting that Ash2L also has a role in chromosome demarcation.
Organizational Affiliation:
Howard Hughes Medical Institute, 1150 West, Medical Center Drive, Ann Arbor, Michigan 48109, USA.