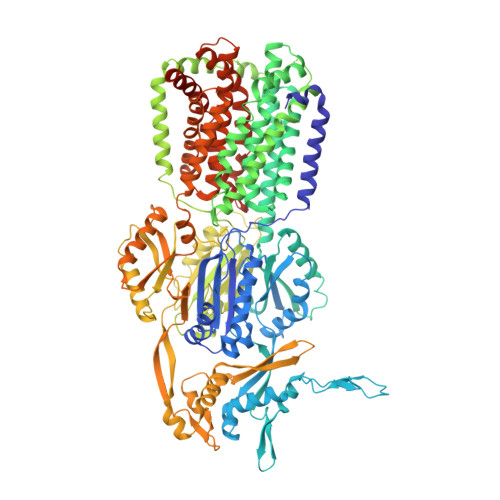
Structure and activity of lipid bilayer within a membrane-protein transporter.
Qiu, W., Fu, Z., Xu, G.G., Grassucci, R.A., Zhang, Y., Frank, J., Hendrickson, W.A., Guo, Y.(2018) Proc Natl Acad Sci U S A 115: 12985-12990
- PubMed: 30509977
- DOI: https://doi.org/10.1073/pnas.1812526115
- Primary Citation of Related Structures:
6BAJ, 6CSX - PubMed Abstract:
Membrane proteins function in native cell membranes, but extraction into isolated particles is needed for many biochemical and structural analyses. Commonly used detergent-extraction methods destroy naturally associated lipid bilayers. Here, we devised a detergent-free method for preparing cell-membrane nanoparticles to study the multidrug exporter AcrB, by cryo-EM at 3.2-Å resolution. We discovered a remarkably well-organized lipid-bilayer structure associated with transmembrane domains of the AcrB trimer. This bilayer patch comprises 24 lipid molecules; inner leaflet chains are packed in a hexagonal array, whereas the outer leaflet has highly irregular but ordered packing. Protein side chains interact with both leaflets and participate in the hexagonal pattern. We suggest that the lipid bilayer supports and harmonizes peristaltic motions through AcrB trimers. In AcrB D407A, a putative proton-relay mutant, lipid bilayer buttresses protein interactions lost in crystal structures after detergent-solubilization. Our detergent-free system preserves lipid-protein interactions for visualization and should be broadly applicable.
Organizational Affiliation:
Department of Medicinal Chemistry, Virginia Commonwealth University, Richmond, VA 23298.