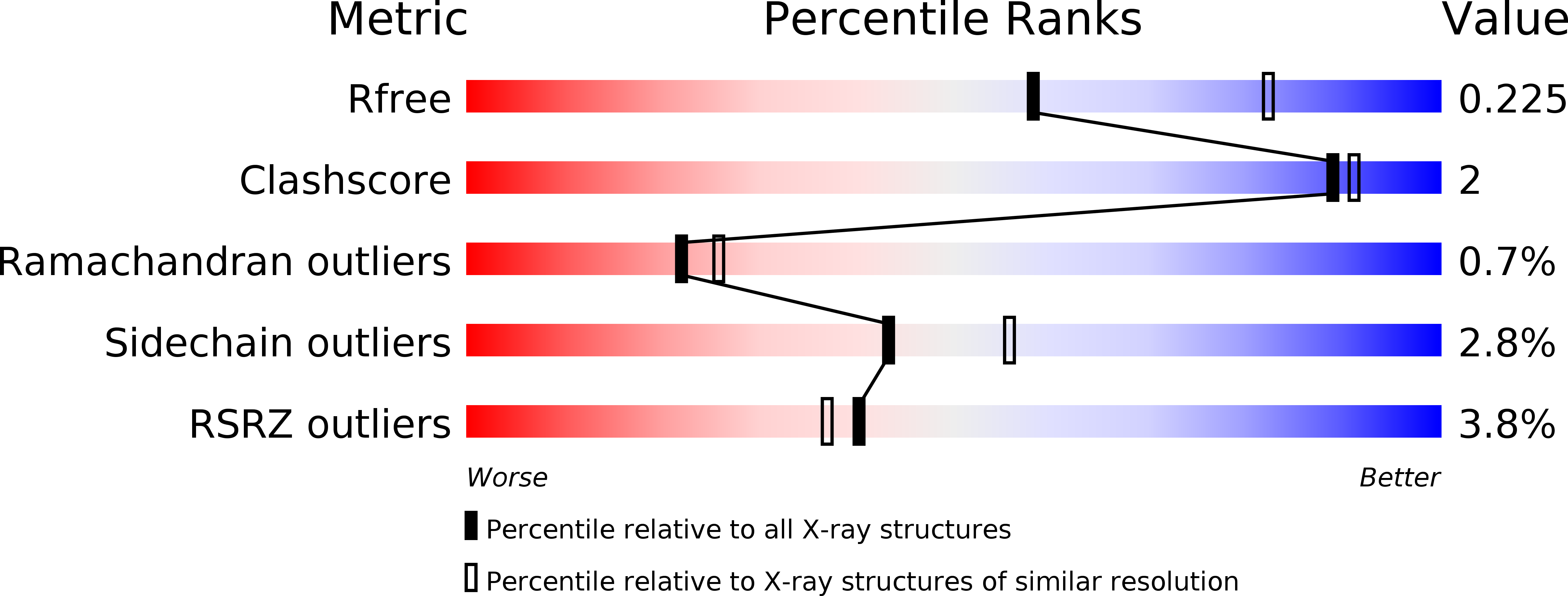
An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Quancard, J., Klein, T., Fung, S.Y., Renatus, M., Hughes, N., Israel, L., Priatel, J.J., Kang, S., Blank, M.A., Viner, R.I., Blank, J., Schlapbach, A., Erbel, P., Kizhakkedathu, J., Villard, F., Hersperger, R., Turvey, S.E., Eder, J., Bornancin, F., Overall, C.M.(2019) Nat Chem Biol 15: 304-313
- PubMed: 30692685
- DOI: https://doi.org/10.1038/s41589-018-0222-1
- Primary Citation of Related Structures:
6F7I, 6H4A - PubMed Abstract:
MALT1 paracaspase is central for lymphocyte antigen-dependent responses including NF-κB activation. We discovered nanomolar, selective allosteric inhibitors of MALT1 that bind by displacing the side chain of Trp580, locking the protease in an inactive conformation. Interestingly, we had previously identified a patient homozygous for a MALT1 Trp580-to-serine mutation who suffered from combined immunodeficiency. We show that the loss of tryptophan weakened interactions between the paracaspase and C-terminal immunoglobulin MALT1 domains resulting in protein instability, reduced protein levels and functions. Upon binding of allosteric inhibitors of increasing potency, we found proportionate increased stabilization of MALT1-W580S to reach that of wild-type MALT1. With restored levels of stable MALT1 protein, the most potent of the allosteric inhibitors rescued NF-κB and JNK signaling in patient lymphocytes. Following compound washout, MALT1 substrate cleavage was partly recovered. Thus, a molecular corrector rescues an enzyme deficiency by substituting for the mutated residue, inspiring new potential precision therapies to increase mutant enzyme activity in other deficiencies.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, Novartis Campus, Basel, Switzerland. jean.quancard@novartis.com.