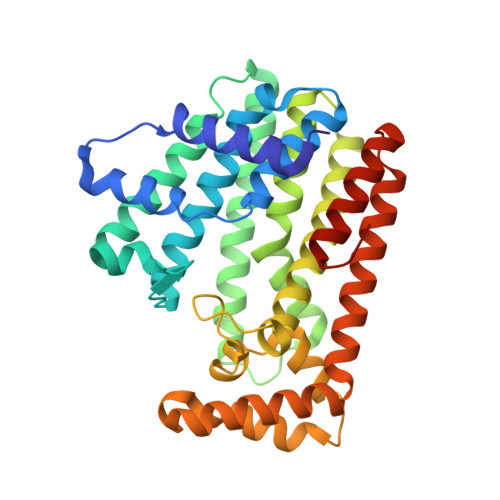
Crystal structure of geranylgeranyl pyrophosphate synthase (crtE) from Nonlabens dokdonensis DSW-6.
Kim, S., Kim, E.J., Park, J.B., Kim, S.W., Kim, K.J.(2019) Biochem Biophys Res Commun 518: 479-485
- PubMed: 31427080
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.071
- Primary Citation of Related Structures:
6KD7 - PubMed Abstract:
Isoprenoids comprise a diverse group of natural products with a broad range of metabolic functions. Isoprenoids are synthesized from prenyl pyrophosphates by prenyltransferases that catalyze the isoprenoid chain-elongation process to different chain lengths. We hereby present the crystal structure of geranylgeranyl pyrophosphate synthase from the marine flavobacterium Nonlabens dokdonensis DSW-6 (NdGGPPS). NdGGPPS forms a hexamer composed of homodimeric trimer, and the monomeric structure is composed of 15 α-helices (α1-α15). In this structure, we observed the binding of one pyrophosphate molecule and two glycerol molecules that mimicked substrate binding to the enzyme. The substrate binding site of NdGGPPS contains large hydrophobic residues such as Phe, His and Tyr, and structural and amino acids sequence analyses thereof suggest that the protein belongs to the short-chain prenyltransferase family.
Organizational Affiliation:
School of Life Sciences, KNU Creative BioResearch Group, Kyungpook National University, Daehak-ro 80, Buk-ku, Daegu, 41566, Republic of Korea; KNU Institute for Microorganisms, Kyungpook National University, Daehak-ro 80, Buk-ku, Daegu 41566, Republic of Korea.