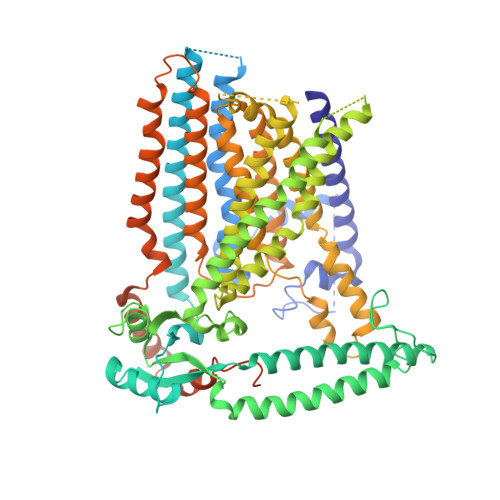
Cryo-EM structure of the mechanically activated ion channel OSCA1.2.
Jojoa Cruz, S., Saotome, K., Murthy, S.E., Tsui, C.C.A., Sansom, M.S., Patapoutian, A., Ward, A.B.(2018) Elife 7
- PubMed: 30382939
- DOI: https://doi.org/10.7554/eLife.41845
- Primary Citation of Related Structures:
6MGV, 6MGW - PubMed Abstract:
Mechanically activated ion channels underlie touch, hearing, shear-stress sensing, and response to turgor pressure. OSCA/TMEM63s are a newly-identified family of eukaryotic mechanically activated ion channels opened by membrane tension. The structural underpinnings of OSCA/TMEM63 function are not explored. Here, we elucidate high resolution cryo-electron microscopy structures of OSCA1.2, revealing a dimeric architecture containing eleven transmembrane helices per subunit and surprising topological similarities to TMEM16 proteins. We locate the ion permeation pathway within each subunit by demonstrating that a conserved acidic residue is a determinant of channel conductance. Molecular dynamics simulations reveal membrane interactions, suggesting the role of lipids in OSCA1.2 gating. These results lay a foundation to decipher how the structural organization of OSCA/TMEM63 is suited for their roles as MA ion channels.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, California, United States.