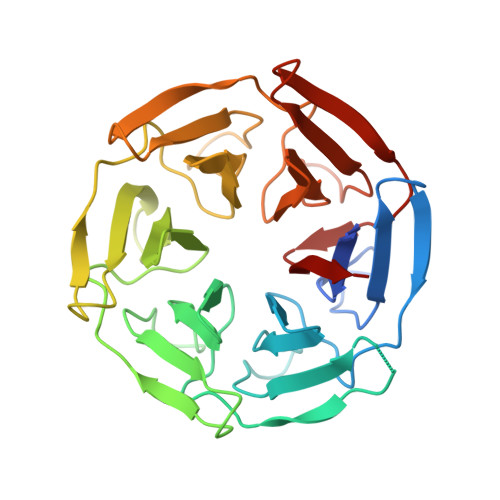
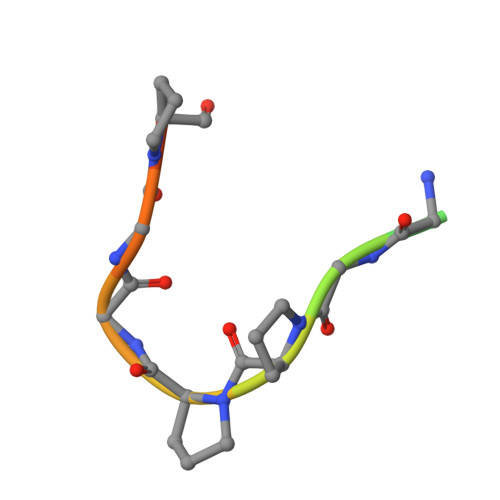
Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Chen, Z., Wasney, G.A., Picaud, S., Filippakopoulos, P., Vedadi, M., D'Angiolella, V., Bullock, A.N.(2020) Open Biol 10: 200041-200041
- PubMed: 32574548
- DOI: https://doi.org/10.1098/rsob.200041
- Primary Citation of Related Structures:
6TTK - PubMed Abstract:
Wnt signalling is dependent on dishevelled proteins (DVL1-3), which assemble an intracellular Wnt signalosome at the plasma membrane. The levels of DVL1-3 are regulated by multiple Cullin-RING E3 ligases that mediate their ubiquitination and degradation. The BTB-Kelch protein KLHL12 was the first E3 ubiquitin ligase to be identified for DVL1-3, but the molecular mechanisms determining its substrate interactions have remained unknown. Here, we mapped the interaction of DVL1-3 to a 'PGXPP' motif that is conserved in other known partners and substrates of KLHL12, including PLEKHA4, PEF1, SEC31 and DRD4. To determine the binding mechanism, we solved a 2.4 Å crystal structure of the Kelch domain of KLHL12 in complex with a DVL1 peptide that bound with low micromolar affinity. The DVL1 substrate adopted a U-shaped turn conformation that enabled hydrophobic interactions with all six blades of the Kelch domain β-propeller. In cells, the mutation or deletion of this motif reduced the binding and ubiquitination of DVL1 and increased its stability confirming this sequence as a degron motif for KLHL12 recruitment. These results define the molecular mechanisms determining DVL regulation by KLHL12 and establish the KLHL12 Kelch domain as a new protein interaction module for a novel proline-rich motif.
Organizational Affiliation:
Structural Genomics Consortium, University of Oxford, Old Road Campus Research Building, Roosevelt Drive, Oxford OX3 7DQ, UK.