Glypicans shield the Wnt lipid moiety to enable signalling at a distance.
McGough, I.J., Vecchia, L., Bishop, B., Malinauskas, T., Beckett, K., Joshi, D., O'Reilly, N., Siebold, C., Jones, E.Y., Vincent, J.P.(2020) Nature 585: 85-90
- PubMed: 32699409
- DOI: https://doi.org/10.1038/s41586-020-2498-z
- Primary Citation of Related Structures:
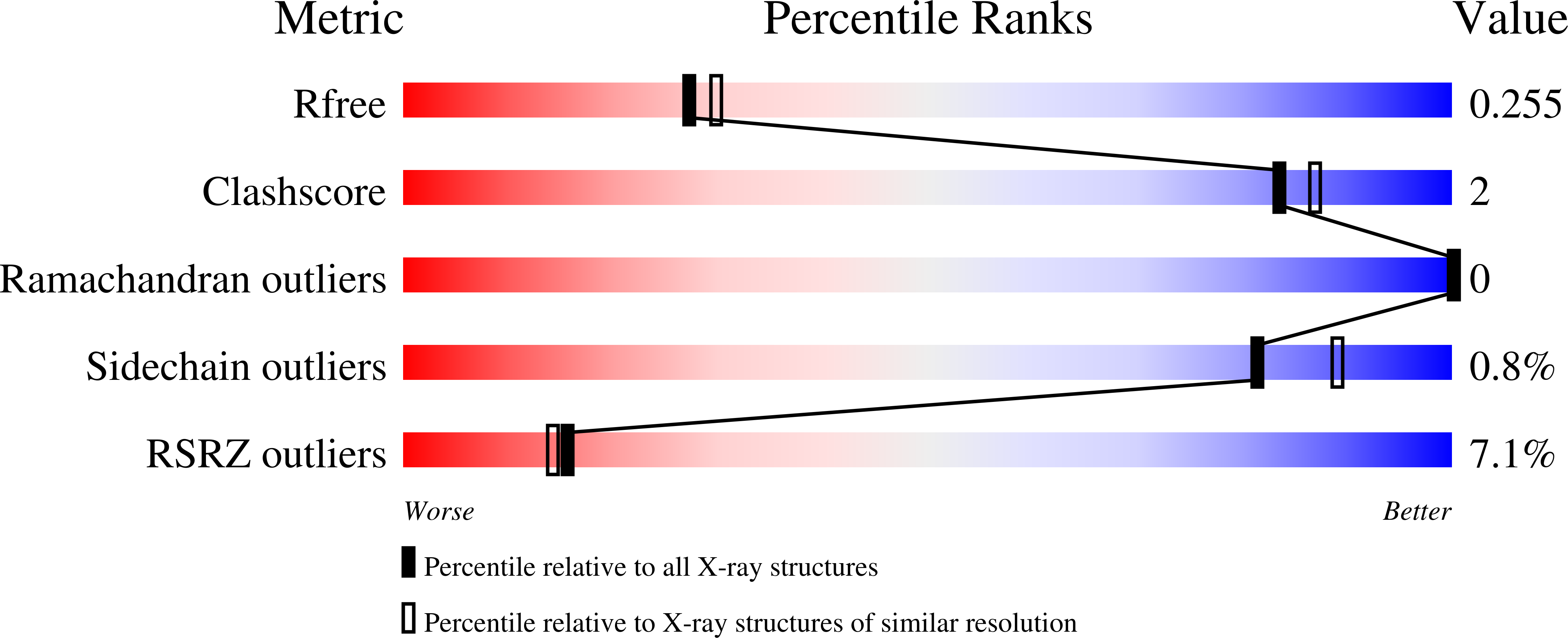
6XTZ - PubMed Abstract:
A relatively small number of proteins have been suggested to act as morphogens-signalling molecules that spread within tissues to organize tissue repair and the specification of cell fate during development. Among them are Wnt proteins, which carry a palmitoleate moiety that is essential for signalling activity 1-3 . How a hydrophobic lipoprotein can spread in the aqueous extracellular space is unknown. Several mechanisms, such as those involving lipoprotein particles, exosomes or a specific chaperone, have been proposed to overcome this so-called Wnt solubility problem 4-6 . Here we provide evidence against these models and show that the Wnt lipid is shielded by the core domain of a subclass of glypicans defined by the Dally-like protein (Dlp). Structural analysis shows that, in the presence of palmitoleoylated peptides, these glypicans change conformation to create a hydrophobic space. Thus, glypicans of the Dlp family protect the lipid of Wnt proteins from the aqueous environment and serve as a reservoir from which Wnt proteins can be handed over to signalling receptors.
Organizational Affiliation:
The Francis Crick Institute, London, UK.