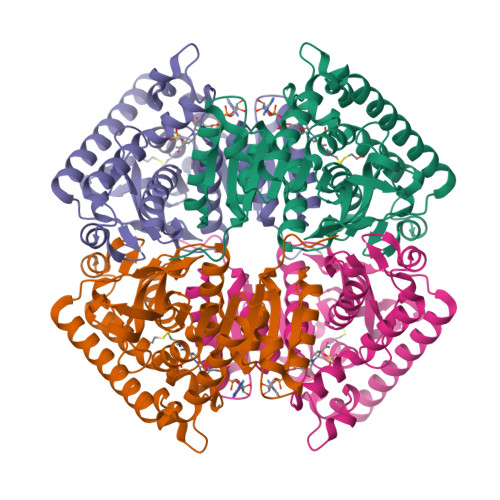
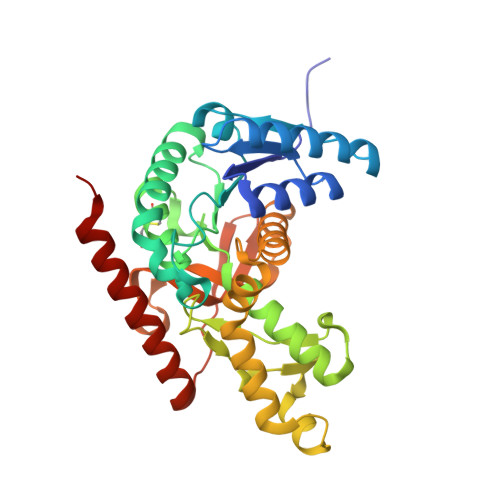
Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Kavanagh, K.L., Elling, R.A., Wilson, D.K.(2004) Biochemistry 43: 879-889
- PubMed: 14744130
- DOI: https://doi.org/10.1021/bi035108g
- Primary Citation of Related Structures:
1PZE, 1PZF, 1PZG, 1PZH - PubMed Abstract:
While within a human host the opportunistic pathogen Toxoplasma gondii relies heavily on glycolysis for its energy needs. Lactate dehydrogenase (LDH), the terminal enzyme in anaerobic glycolysis necessary for NAD(+) regeneration, therefore represents an attractive therapeutic target. The tachyzoite stage lactate dehydrogenase (LDH1) from the parasite T. gondii has been crystallized in apo form and in ternary complexes containing NAD(+) or the NAD(+)-analogue 3-acetylpyridine adenine dinucleotide (APAD(+)) and sulfate or the inhibitor oxalate. Comparison of the apo and ternary models shows an active-site loop that becomes ordered upon substrate binding. This active-site loop is five residues longer than in most LDHs and necessarily adopts a different conformation. While loop isomerization is fully rate-limiting in prototypical LDHs, kinetic data suggest that LDH1's rate is limited by chemical steps. The importance of charge neutralization in ligand binding is supported by the complexes that have been crystallized as well as fluorescence quenching experiments performed with ligands at low and high pH. A methionine that replaces a serine residue and displaces an ordered water molecule often seen in LDH structures provides a structural explanation for reduced substrate inhibition. Superimposition of LDH1 with human muscle- and heart-specific LDH isoforms reveals differences in residues that line the active site that increase LDH1's hydrophobicity. These differences will aid in designing inhibitors specific for LDH1 that may be useful in treating toxoplasmic encephalitis and other complications that arise in immune-compromised individuals.
Organizational Affiliation:
Section of Molecular and Cellular Biology, University of California, Davis, California 95616, USA.