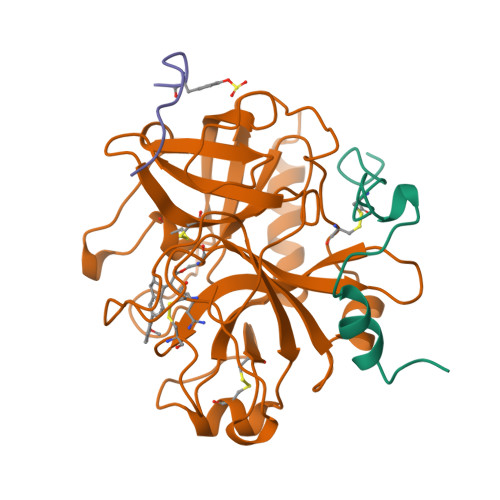
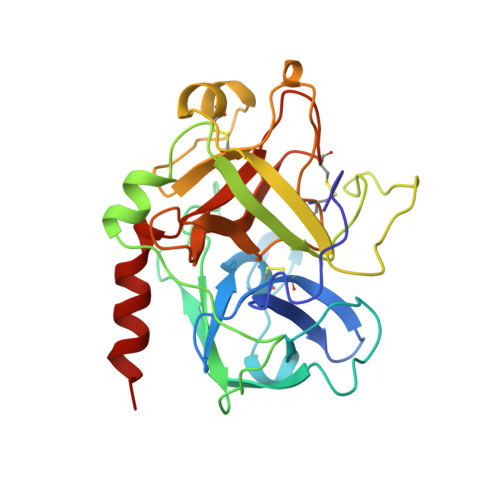
Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Jhoti, H., Cleasby, A., Reid, S., Thomas, P., Weir, M., Wonacott, A.(1999) Biochemistry 38: 7969
- PubMed: 10387040
- DOI: https://doi.org/10.1021/bi9830359
- Primary Citation of Related Structures:
1QHR, 1QJ1, 1QJ6, 1QJ7 - PubMed Abstract:
The binding modes of four active site-directed, acylating inhibitors of human alpha-thrombin have been determined using X-ray crystallography. These inhibitors (GR157368, GR166081, GR167088, and GR179849) are representatives of a series utilizing a novel 5, 5-trans-lactone template to specifically acylate Ser195 of thrombin, resulting in an acyl complex. In each case the crystal structure of the complex reveals a binding mode which is consistent with the formation of a covalent bond between the ring-opened lactone of the inhibitor and residue Ser195. Improvements in potency and selectivity of these inhibitors for thrombin are rationalized on the basis of the observed protein/inhibitor interactions identified in these complexes. Occupation of the thrombin S2 and S3 pockets is shown to be directly correlated with improved binding and a degree of selectivity. The binding mode of GR179849 to thrombin is compared with the thrombin/PPACK complex [Bode, W., Turk, D., and Karshikov, A. (1992) Protein Sci. 1, 426-471] as this represents the archetypal binding mode for a thrombin inhibitor. This series of crystal structures is the first to be reported of synthetic, nonpeptidic acylating inhibitors bound to thrombin and provides details of the molecular recognition features that resulted in nanomolar potency.
Organizational Affiliation:
Biomolecular Structure Unit, Glaxo Wellcome Medicines Research Centre, Hertfordshire, U.K.