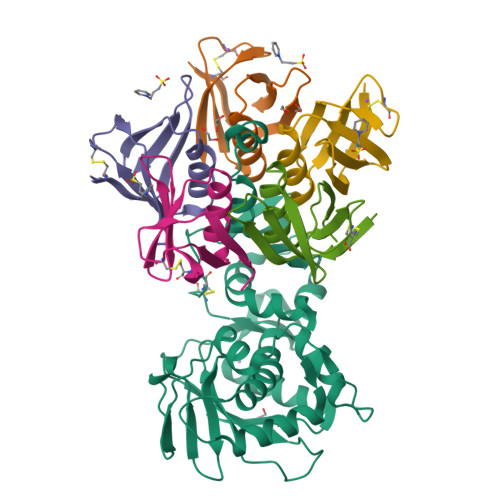
Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
Fraser, M.E., Fujinaga, M., Cherney, M.M., Melton-Celsa, A.R., Twiddy, E.M., O'Brien, A.D., James, M.N.G.(2004) J Biological Chem 279: 27511-27517
- PubMed: 15075327
- DOI: https://doi.org/10.1074/jbc.M401939200
- Primary Citation of Related Structures:
1R4P, 1R4Q - PubMed Abstract:
Several serotypes of Escherichia coli produce protein toxins closely related to Shiga toxin (Stx) from Shigella dysenteriae serotype 1. These Stx-producing E. coli cause outbreaks of hemorrhagic colitis and hemolytic uremic syndrome in humans, with the latter being more likely if the E. coli produce Stx2 than if they only produce Stx1. To investigate the differences among the Stxs, which are all AB(5) toxins, the crystal structure of Stx2 from E. coli O157:H7 was determined at 1.8-A resolution and compared with the known structure of Stx. Our major finding was that, in contrast to Stx, the active site of the A-subunit of Stx2 is accessible in the holotoxin, and a molecule of formic acid and a water molecule mimic the binding of the adenine base of the substrate. Further, the A-subunit adopts a different orientation with respect to the B-subunits in Stx2 than in Stx, due to interactions between the carboxyl termini of the B-subunits and neighboring regions of the A-subunit. Of the three types of receptor-binding sites in the B-pentamer, one has a different conformation in Stx2 than in Stx, and the carboxyl terminus of the A-subunit binds at another. Any of these structural differences might result in different mechanisms of action of the two toxins and the development of hemolytic uremic syndrome upon exposure to Stx2.
Organizational Affiliation:
Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada.