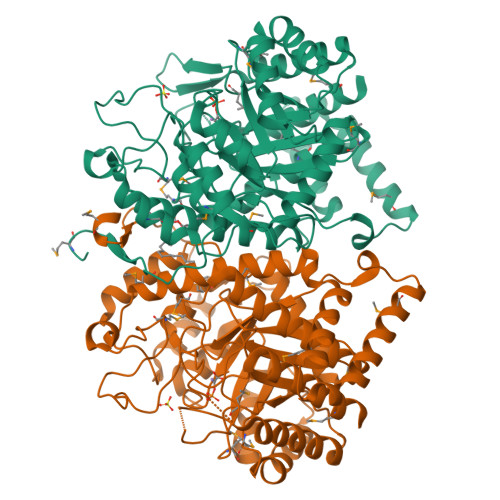
The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis reveals a common catalytic scaffold and ancestry for type I and type II enzymes
Webby, C.J., Baker, H.M., Lott, J.S., Baker, E.N., Parker, E.J.(2005) J Mol Biology 354: 927-939
- PubMed: 16288916
- DOI: https://doi.org/10.1016/j.jmb.2005.09.093
- Primary Citation of Related Structures:
2B7O - PubMed Abstract:
The shikimate pathway, responsible for the biosynthesis of aromatic compounds, is essential for the growth of Mycobacterium tuberculosis and is a potential target for the design of new anti-tuberculosis drugs. The first step of this pathway is catalyzed by 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS). The DAH7PSs have been classified into two apparently unrelated types and, whereas structural data have been obtained for the type I DAH7PSs, no structural information is available for their type II counterparts. The type II DAH7PS from M.tuberculosis has been expressed in Escherichia coli, purified, functionally characterized and crystallized. It is found to be metal ion-dependent and subject to feedback inhibition by phenylalanine, tryptophan, tyrosine and chorismate, with a significant synergistic effect when tryptophan is used in combination with phenylalanine. The crystal structure of M.tuberculosis DAH7PS has been determined by single-wavelength anomalous diffraction and refined at 2.3A in complex with substrate phosphoenolpyruvate and Mn(2+). The structure reveals a tightly associated dimer of (beta/alpha)(8) TIM barrels. The monomer fold, the arrangement of key residues in the active site, and the binding modes of PEP and Mn(2+), all match those of the type I enzymes, and indicate a common ancestry for the type I and type II DAH7PSs, despite their minimal sequence identity. In contrast, the structural elements that decorate the core (beta/alpha)(8) fold differ from those in the type I enzymes, consistent with their different regulatory and oligomeric properties.
Organizational Affiliation:
Centre of Structural Biology, Institute of Fundamental Sciences, Massey University, Palmerston North, New Zealand.