Structure, Modifications and Ligand-Binding Properties of Rat Profilin 2A.
Haikarainen, T., Chen, W.Q., Lubec, G., Kursula, P.(2009) Acta Crystallogr D Biol Crystallogr 65: 303
- PubMed: 19307711
- DOI: https://doi.org/10.1107/S0907444909000699
- Primary Citation of Related Structures:
2VK3 - PubMed Abstract:
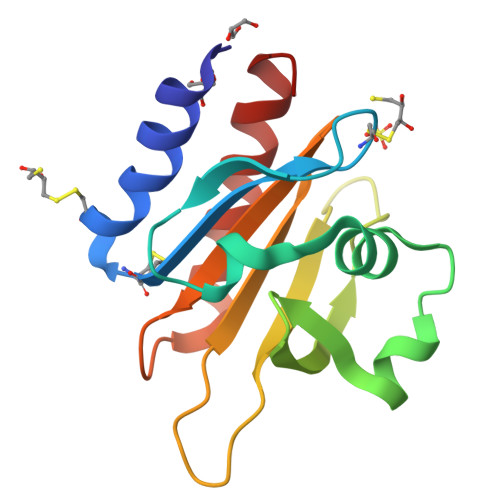
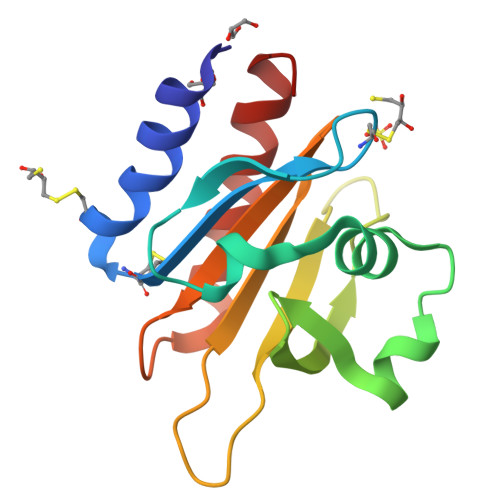
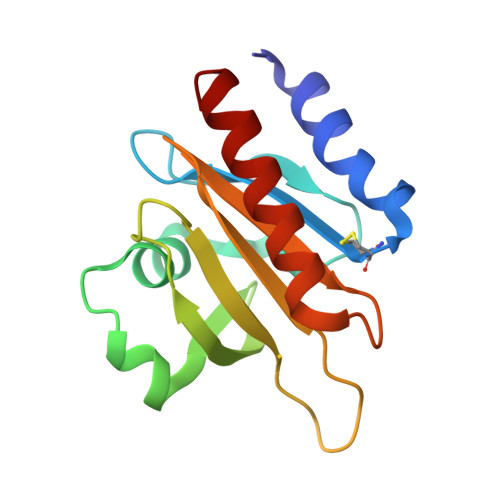
Profilins are key regulators of the actin microfilament system and in neuronal tissues the profilin 2a isoform is the most abundant and important profilin. The high-resolution crystal structure of rat profilin 2a has been determined in the absence of ligands. By comparing the structure with those of peptide-liganded profilin 2a and unliganded profilin 2b, it can be concluded that the binding site for proline-rich peptides is pre-organized. The C-terminus of profilin 2a is also well ordered in the absence of ligand peptide, in contrast to the 2b isoform which is generated by alternative splicing. Covalent modifications of four cysteine residues were also detected in profilin 2a, as well as a number of other modifications in profilin 2 from rat brain; such modifications could significantly affect the function of profilin. It was also shown that profilin 2a binds to the neuronal protein palladin, including a synthetic palladin peptide; peptides from another profilin ligand, dynamin 1, failed to interact with both profilin 1 and profilin 2a. These results allow a better understanding of the structure-function relationships and ligand binding of mammalian profilin 2a.
Organizational Affiliation:
Department of Biochemistry, University of Oulu, Oulu, Finland.