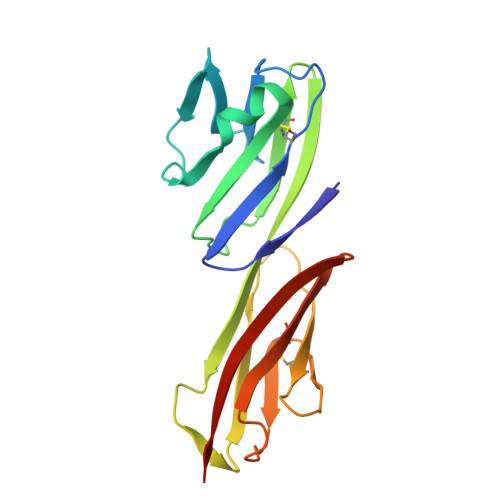
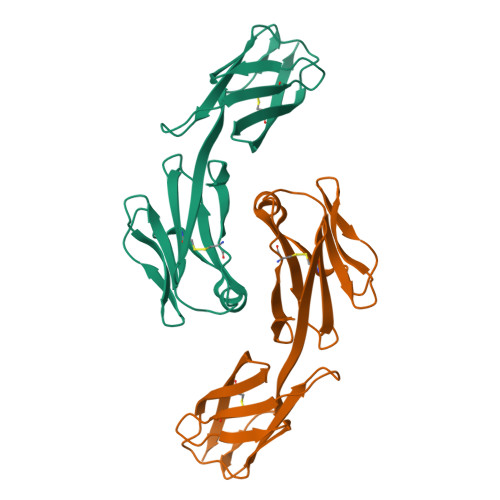
Refinement and analysis of the structure of the first two domains of human CD4
Garrett, T.P.J., Wang, J., Yan, Y., Liu, J., Harrison, S.C.(1993) J Mol Biology 234: 763-778
- PubMed: 8254672
- DOI: https://doi.org/10.1006/jmbi.1993.1625
- Primary Citation of Related Structures:
3CD4 - PubMed Abstract:
The structure of a fragment of human CD4 containing two immunoglobulin (Ig)-like domains has been determined by X-ray crystallography and refined at 2.2 A resolution. The structure determination involved iterative building and simulated-annealing refinement, beginning with a partial model. Comparison of domain 1 with an Ig variable domain shows that CD4 has a long and prominent CDR2-like loop (the C"C" corner) and shortened CC' and FG loops (which mediate dimerization in IgV modules). Comparison of domain 2 with Ig modules and domain 1 shows that it can be described as a truncated Ig V domain, in which strands C" and D are deleted. The intersheet disulfide in domain 2 is absent, and there is an altered packing of the two beta-sheets together with a remodeling of the hydrophobic core. The interface between domains 1 and 2 is a lap joint with an extensive hydrophobic surface. The key features of domain 1 that contribute to the interface are found at corresponding positions in domain 2, leading us to propose that the contact between domains 2 and 3 will resemble the one between domains 1 and 2.
Organizational Affiliation:
Howard Hughes Medical Institute, Cambridge, MA.