Structural Determinants of MALT1 Protease Activity.
Wiesmann, C., Leder, L., Blank, J., Bernardi, A., Melkko, S., Decock, A., D'Arcy, A., Villard, F., Erbel, P., Hughes, N., Freuler, F., Nikolay, R., Alves, J., Bornancin, F., Renatus, M.(2012) J Mol Biology 419: 4-21
- PubMed: 22366302
- DOI: https://doi.org/10.1016/j.jmb.2012.02.018
- Primary Citation of Related Structures:
3V4L, 3V4O, 3V55 - PubMed Abstract:
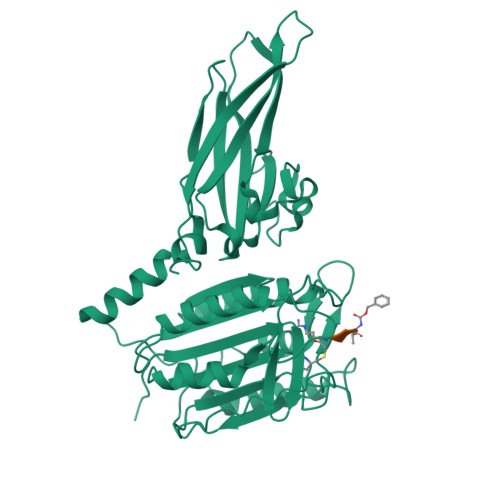
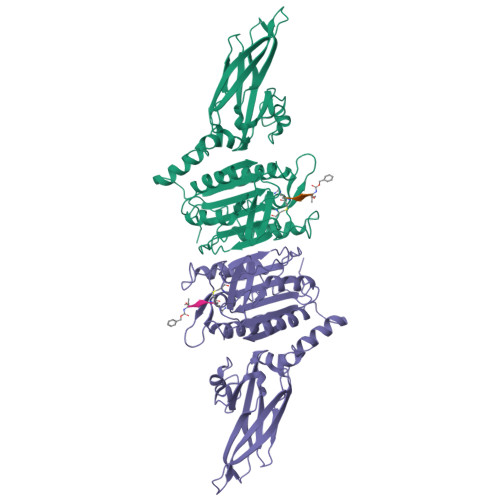
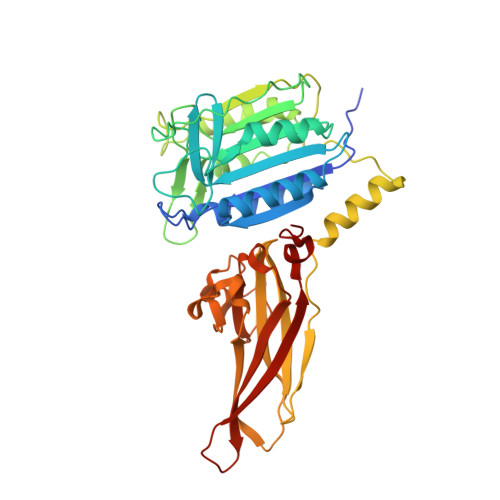
The formation of the CBM (CARD11-BCL10-MALT1) complex is pivotal for antigen-receptor-mediated activation of the transcription factor NF-κB. Signaling is dependent on MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1), which not only acts as a scaffolding protein but also possesses proteolytic activity mediated by its caspase-like domain. It remained unclear how the CBM activates MALT1. Here, we provide biochemical and structural evidence that MALT1 activation is dependent on its dimerization and show that mutations at the dimer interface abrogate activity in cells. The unliganded protease presents itself in a dimeric yet inactive state and undergoes substantial conformational changes upon substrate binding. These structural changes also affect the conformation of the C-terminal Ig-like domain, a domain that is required for MALT1 activity. Binding to the active site is coupled to a relative movement of caspase and Ig-like domains. MALT1 binding partners thus may have the potential of tuning MALT1 protease activity without binding directly to the caspase domain.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, Forum 1, Novartis Campus, CH-4002 Basel, Switzerland.