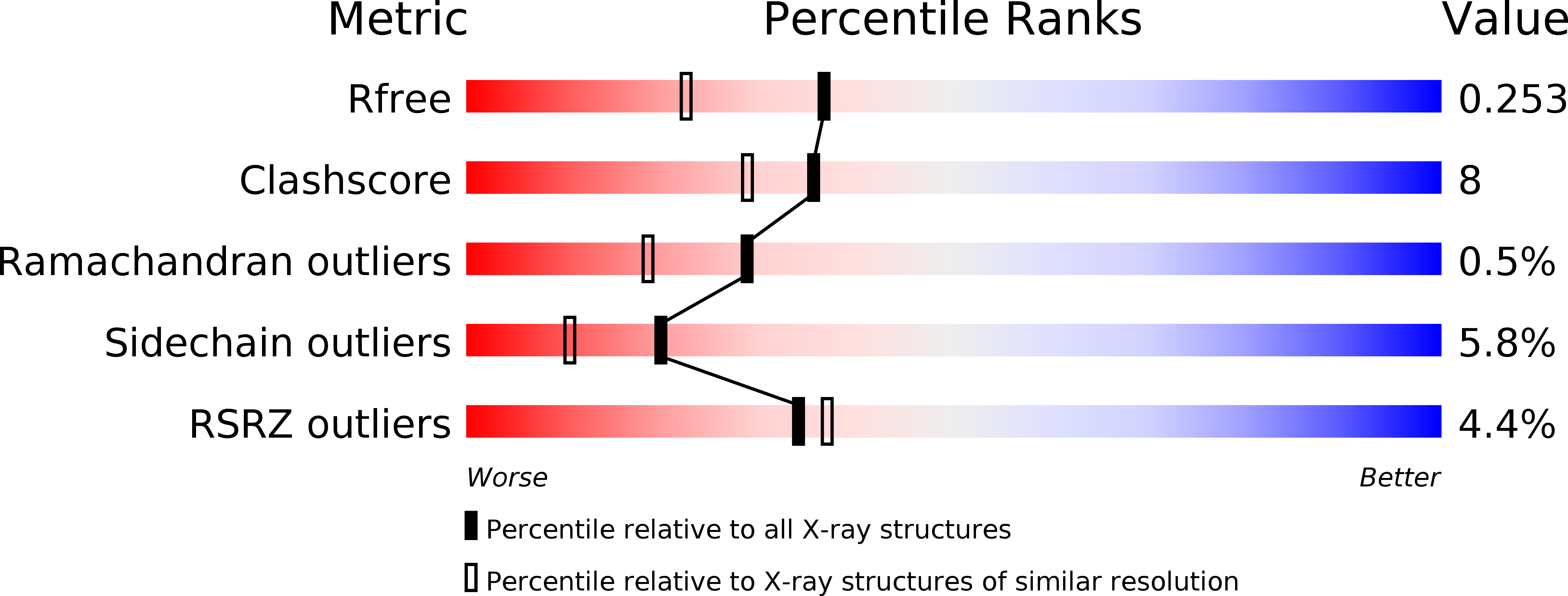
Mutational and crystallographic analysis of l-amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813: Interconversion between oxidase and monooxygenase activities
Matsui, D., Im, D.H., Sugawara, A., Fukuta, Y., Fushinobu, S., Isobe, K., Asano, Y.(2014) FEBS Open Bio 4: 220-228
- PubMed: 24693490
- DOI: https://doi.org/10.1016/j.fob.2014.02.002
- Primary Citation of Related Structures:
3WE0 - PubMed Abstract:
In this study, it was shown for the first time that l-amino acid oxidase of Pseudomonas sp. AIU813, renamed as l-amino acid oxidase/monooxygenase (l-AAO/MOG), exhibits l-lysine 2-monooxygenase as well as oxidase activity. l-Lysine oxidase activity of l-AAO/MOG was increased in a p-chloromercuribenzoate (p-CMB) concentration-dependent manner to a final level that was five fold higher than that of the non-treated enzyme. In order to explain the effects of modification by the sulfhydryl reagent, saturation mutagenesis studies were carried out on five cysteine residues, and we succeeded in identifying l-AAO/MOG C254I mutant enzyme, which showed five-times higher specific activity of oxidase activity than that of wild type. The monooxygenase activity shown by the C254I variant was decreased significantly. Moreover, we also determined a high-resolution three-dimensional structure of l-AAO/MOG to provide a structural basis for its biochemical characteristics. The key residue for the activity conversion of l-AAO/MOG, Cys-254, is located near the aromatic cage (Trp-418, Phe-473, and Trp-516). Although the location of Cys-254 indicates that it is not directly involved in the substrate binding, the chemical modification by p-CMB or C254I mutation would have a significant impact on the substrate binding via the side chain of Trp-516. It is suggested that a slight difference of the binding position of a substrate can dictate the activity of this type of enzyme as oxidase or monooxygenase.
Organizational Affiliation:
Biotechnology Research Center and Department of Biotechnology, Toyama Prefectural University, 5180 Kurokawa, Imizu, Toyama 939-0398, Japan ; Asano Active Enzyme Molecule Project, ERATO, JST, 5180 Kurokawa, Imizu,Toyama 939-0398, Japan.