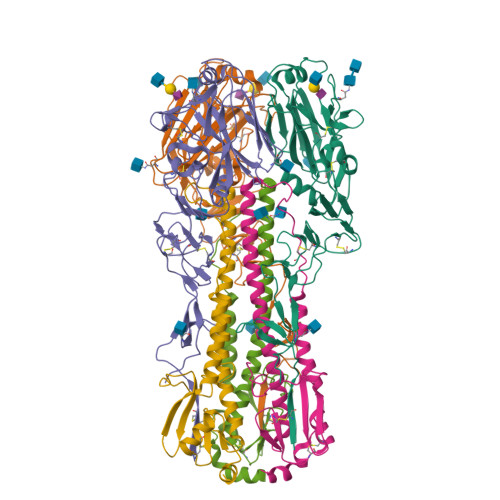
Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus.
Yang, H., Carney, P.J., Donis, R.O., Stevens, J.(2012) J Virol 86: 8645-8652
- PubMed: 22674977
- DOI: https://doi.org/10.1128/JVI.00281-12
- Primary Citation of Related Structures:
4DJ6, 4DJ7, 4DJ8 - PubMed Abstract:
Recurrence of highly pathogenic avian influenza (HPAI) virus subtype H7 in poultry continues to be a public health concern. In 2003, an HPAI H7N7 outbreak in The Netherlands infected 89 people in close contact with affected poultry and resulted in one fatal case. In previous studies, the virus isolated from this fatal case, A/Netherlands/219/2003 (NL219) caused a lethal infection in mouse models and had increased replication efficiency and a broader tissue distribution than nonlethal isolates from the same outbreak. A mutation which introduces a potential glycosylation site at Asn123 in the NL219 hemagglutinin was postulated to contribute to the pathogenic properties of this virus. To study this further, we have expressed the NL219 hemagglutinin in a baculovirus expression system and performed a structural analysis of the hemagglutinin in complex with avian and human receptor analogs. Glycan microarray and kinetic analysis were performed to compare the receptor binding profile of the wild-type recombinant NL219 HA to a variant with a threonine-to-alanine mutation at position 125, resulting in loss of the glycosylation site at Asn123. The results suggest that the additional glycosylation sequon increases binding affinity to avian-type α2-3-linked sialosides rather than switching to a human-like receptor specificity and highlight the mechanistic diversity of these pathogens, which calls attention to the need for further studies to fully understand the unique properties of these viruses.
Organizational Affiliation:
Influenza Division, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.