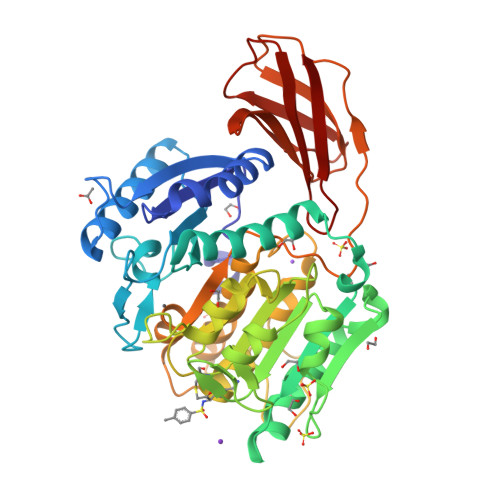
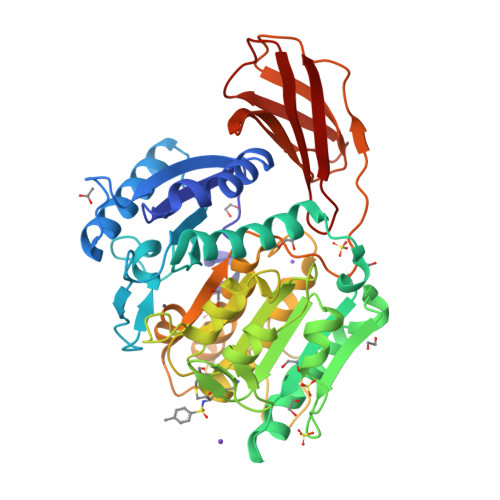
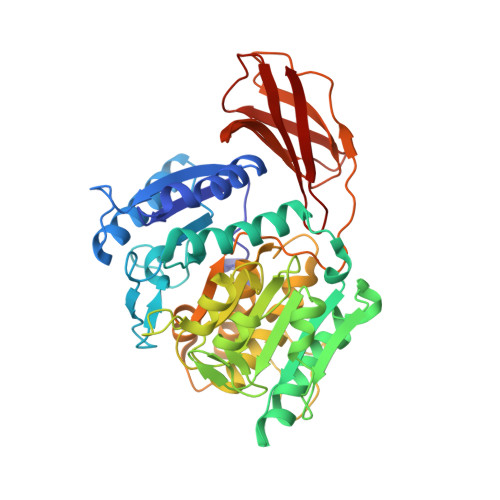
Structure of the lysine specific protease Kgp from Porphyromonas gingivalis, a target for improved oral health.
Gorman, M.A., Seers, C.A., Michell, B.J., Feil, S.C., Huq, N.L., Cross, K.J., Reynolds, E.C., Parker, M.W.(2015) Protein Sci 24: 162-166
- PubMed: 25327141
- DOI: https://doi.org/10.1002/pro.2589
- Primary Citation of Related Structures:
4TKX - PubMed Abstract:
The oral pathogen Porphyromonas gingivalis is a keystone pathogen in the development of chronic periodontitis. Gingipains, the principle virulence factors of P. gingivalis are multidomain, cell-surface proteins containing a cysteine protease domain. The lysine specific gingipain, Kgp, is a critical virulence factor of P. gingivalis. We have determined the X-ray crystal structure of the lysine-specific protease domain of Kgp to 1.6 Å resolution. The structure provides insights into the mechanism of substrate specificity and catalysis.
Organizational Affiliation:
ACRF Rational Drug Discovery Centre, St. Vincent's Institute of Medical Research, Fitzroy, Victoria, 3065, Australia.