Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Reich, S., Guilligay, D., Pflug, A., Malet, H., Berger, I., Crepin, T., Hart, D., Lunardi, T., Nanao, M., Ruigrok, R.W., Cusack, S.(2014) Nature 516: 361-366
- PubMed: 25409151
- DOI: https://doi.org/10.1038/nature14009
- Primary Citation of Related Structures:
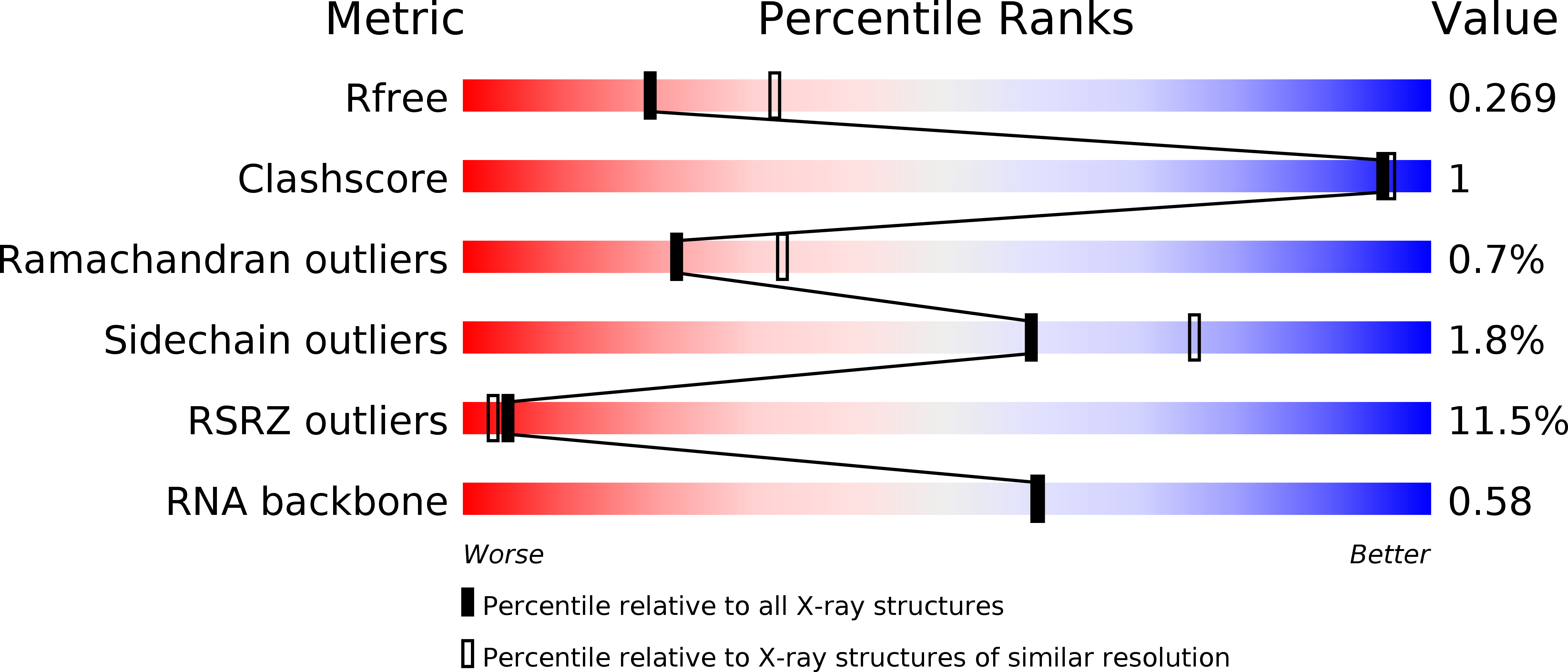
4WRT, 4WSA, 4WSB - PubMed Abstract:
Influenza virus polymerase uses a capped primer, derived by 'cap-snatching' from host pre-messenger RNA, to transcribe its RNA genome into mRNA and a stuttering mechanism to generate the poly(A) tail. By contrast, genome replication is unprimed and generates exact full-length copies of the template. Here we use crystal structures of bat influenza A and human influenza B polymerases (FluA and FluB), bound to the viral RNA promoter, to give mechanistic insight into these distinct processes. In the FluA structure, a loop analogous to the priming loop of flavivirus polymerases suggests that influenza could initiate unprimed template replication by a similar mechanism. Comparing the FluA and FluB structures suggests that cap-snatching involves in situ rotation of the PB2 cap-binding domain to direct the capped primer first towards the endonuclease and then into the polymerase active site. The polymerase probably undergoes considerable conformational changes to convert the observed pre-initiation state into the active initiation and elongation states.
Organizational Affiliation:
1] European Molecular Biology Laboratory, Grenoble Outstation, 71 Avenue des Martyrs, CS 90181, 38042 Grenoble Cedex 9, France [2] University Grenoble Alpes-Centre National de la Recherche Scientifique-EMBL Unit of Virus Host-Cell Interactions, 71 Avenue des Martyrs, CS 90181, 38042 Grenoble Cedex 9, France.