Insights into Autoregulation of Notch3 from Structural and Functional Studies of Its Negative Regulatory Region.
Xu, X., Choi, S.H., Hu, T., Tiyanont, K., Habets, R., Groot, A.J., Vooijs, M., Aster, J.C., Chopra, R., Fryer, C., Blacklow, S.C.(2015) Structure 23: 1227-1235
- PubMed: 26051713
- DOI: https://doi.org/10.1016/j.str.2015.05.001
- Primary Citation of Related Structures:
4ZLP - PubMed Abstract:
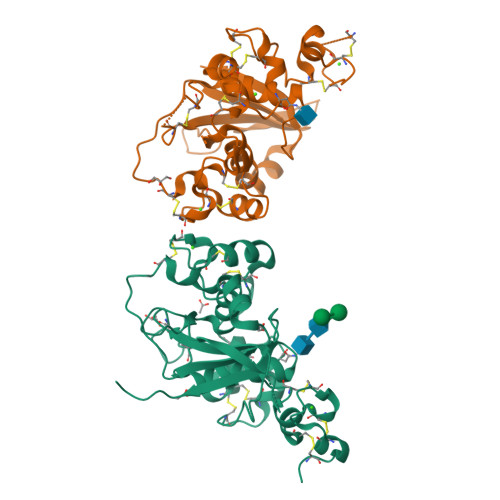
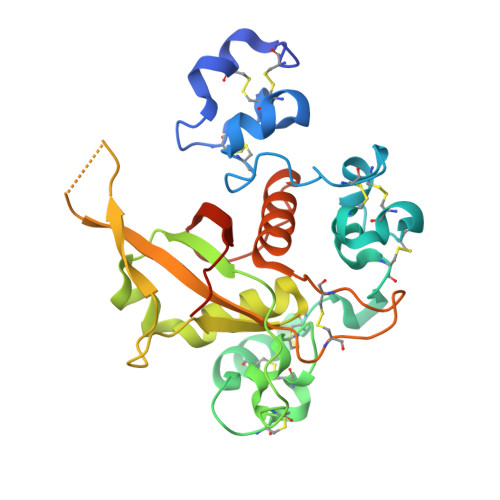
Notch receptors are transmembrane proteins that undergo activating proteolysis in response to ligand stimulation. A negative regulatory region (NRR) maintains receptor quiescence by preventing protease cleavage prior to ligand binding. We report here the X-ray structure of the NRR of autoinhibited human Notch3, and compare it with the Notch1 and Notch2 NRRs. The overall architecture of the autoinhibited conformation, in which three LIN12-Notch repeat (LNR) modules wrap around a heterodimerization domain, is preserved in Notch3, but the autoinhibited conformation of the Notch3 NRR is less stable. The Notch3 NRR uses a highly conserved surface on the third LNR module to form a dimer in the crystal. Similar homotypic interfaces exist in Notch1 and Notch2. Together, these studies reveal distinguishing structural features associated with increased basal activity of Notch3, demonstrate increased ligand-independent signaling for disease-associated mutations that map to the Notch3 NRR, and identify a conserved dimerization interface present in multiple Notch receptors.
Organizational Affiliation:
Department of Cancer Biology, Dana Farber Cancer Institute, Boston, MA 02215, USA; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.