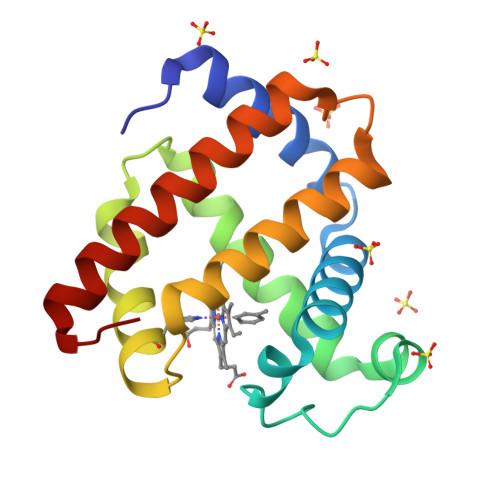
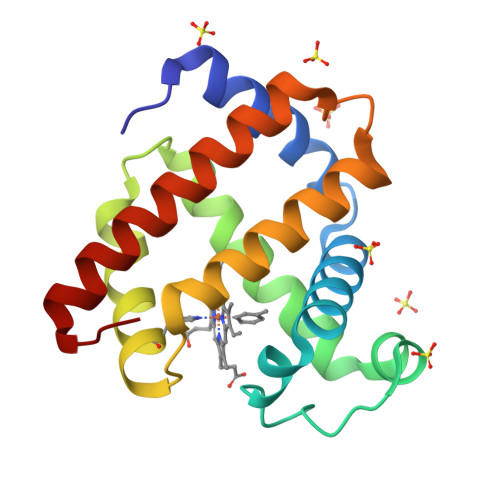
Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
Wang, B., Thomas, L.M., Richter-Addo, G.B.(2016) J Inorg Biochem 164: 1-4
- PubMed: 27687333
- DOI: https://doi.org/10.1016/j.jinorgbio.2016.06.028
- Primary Citation of Related Structures:
5IKS, 5ILE, 5ILM, 5ILP, 5ILR - PubMed Abstract:
Bioorganometallic Fe-C bonds are biologically relevant species that may result from the metabolism of natural or synthetic hydrazines. The molecular structures of four new sperm whale mutant myoglobin derivatives with Fe-aryl moieties, namely H64A-tolyl-m, H64A-chlorophenyl-p, H64Q-tolyl-m, and H64Q-chlorophenyl-p, have been determined at 1.7-1.9Å resolution. The structures reveal conformational preferences for the substituted aryls resulting from attachment of the aryl ligands to Fe at the site of net -NHNH 2 release from the precursor hydrazines, and show distal pocket changes that readily accommodate these bulky ligands.
Organizational Affiliation:
Department of Chemistry and Biochemistry, and Price Family Foundation Institute of Structural Biology, University of Oklahoma, Norman 73019, United States.