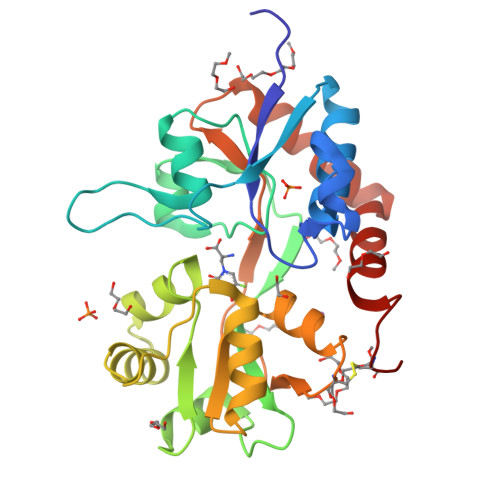
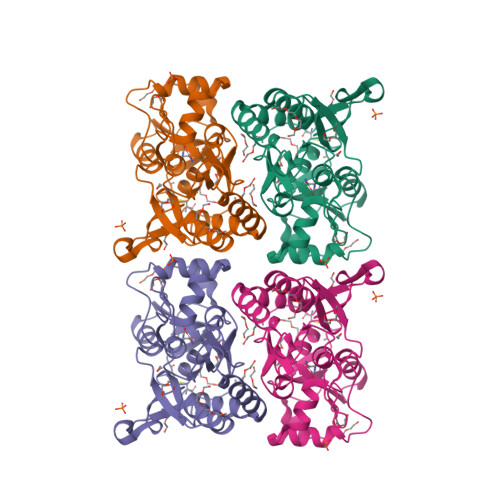
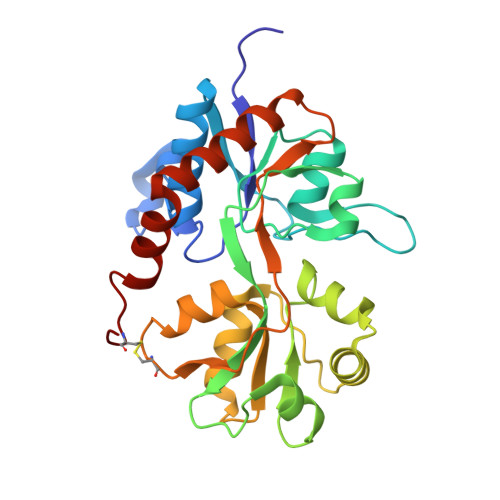
Mechanism of partial agonism in AMPA-type glutamate receptors.
Salazar, H., Eibl, C., Chebli, M., Plested, A.(2017) Nat Commun 8: 14327-14327
- PubMed: 28211453
- DOI: https://doi.org/10.1038/ncomms14327
- Primary Citation of Related Structures:
5JEI - PubMed Abstract:
Neurotransmitters trigger synaptic currents by activating ligand-gated ion channel receptors. Whereas most neurotransmitters are efficacious agonists, molecules that activate receptors more weakly-partial agonists-also exist. Whether these partial agonists have weak activity because they stabilize less active forms, sustain active states for a lesser fraction of the time or both, remains an open question. Here we describe the crystal structure of an α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor (AMPAR) ligand binding domain (LBD) tetramer in complex with the partial agonist 5-fluorowillardiine (FW). We validate this structure, and others of different geometry, using engineered intersubunit bridges. We establish an inverse relation between the efficacy of an agonist and its promiscuity to drive the LBD layer into different conformations. These results suggest that partial agonists of the AMPAR are weak activators of the receptor because they stabilize multiple non-conducting conformations, indicating that agonism is a function of both the space and time domains.
Organizational Affiliation:
Leibniz-Institut für Molekulare Pharmakologie, Robert-Rössle-Strasse 10, 13125 Berlin, Germany.