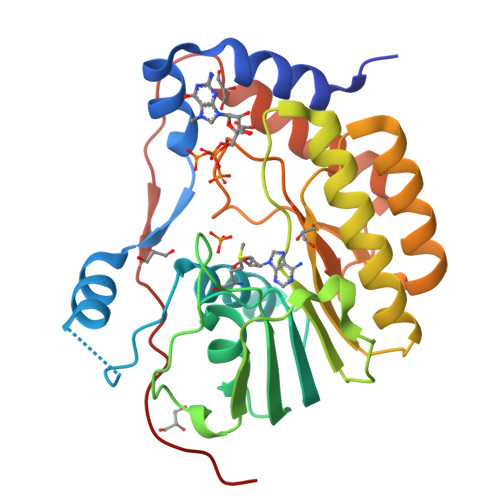
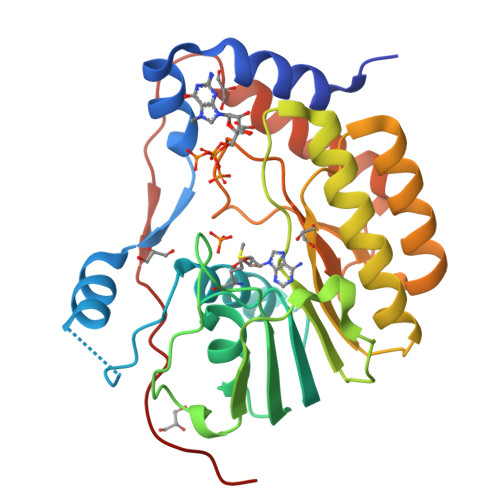
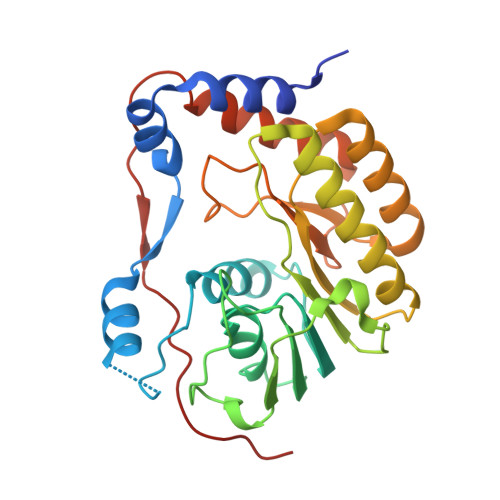
Structures of NS5 Methyltransferase from Zika Virus.
Coloma, J., Jain, R., Rajashankar, K.R., Garcia-Sastre, A., Aggarwal, A.K.(2016) Cell Rep 16: 3097-3102
- PubMed: 27633330
- DOI: https://doi.org/10.1016/j.celrep.2016.08.091
- Primary Citation of Related Structures:
5KQR, 5KQS - PubMed Abstract:
The Zika virus (ZIKV) poses a major public health emergency. To aid in the development of antivirals, we present two high-resolution crystal structures of the ZIKV NS5 methyltransferase: one bound to S-adenosylmethionine (SAM) and the other bound to SAM and 7-methyl guanosine diphosphate (7-MeGpp). We identify features of ZIKV NS5 methyltransferase that lend to structure-based antiviral drug discovery. Specifically, SAM analogs with functionalities on the Cβ atom of the methionine portion of the molecules that occupy the RNA binding tunnel may provide better specificity relative to human RNA methyltransferases.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.