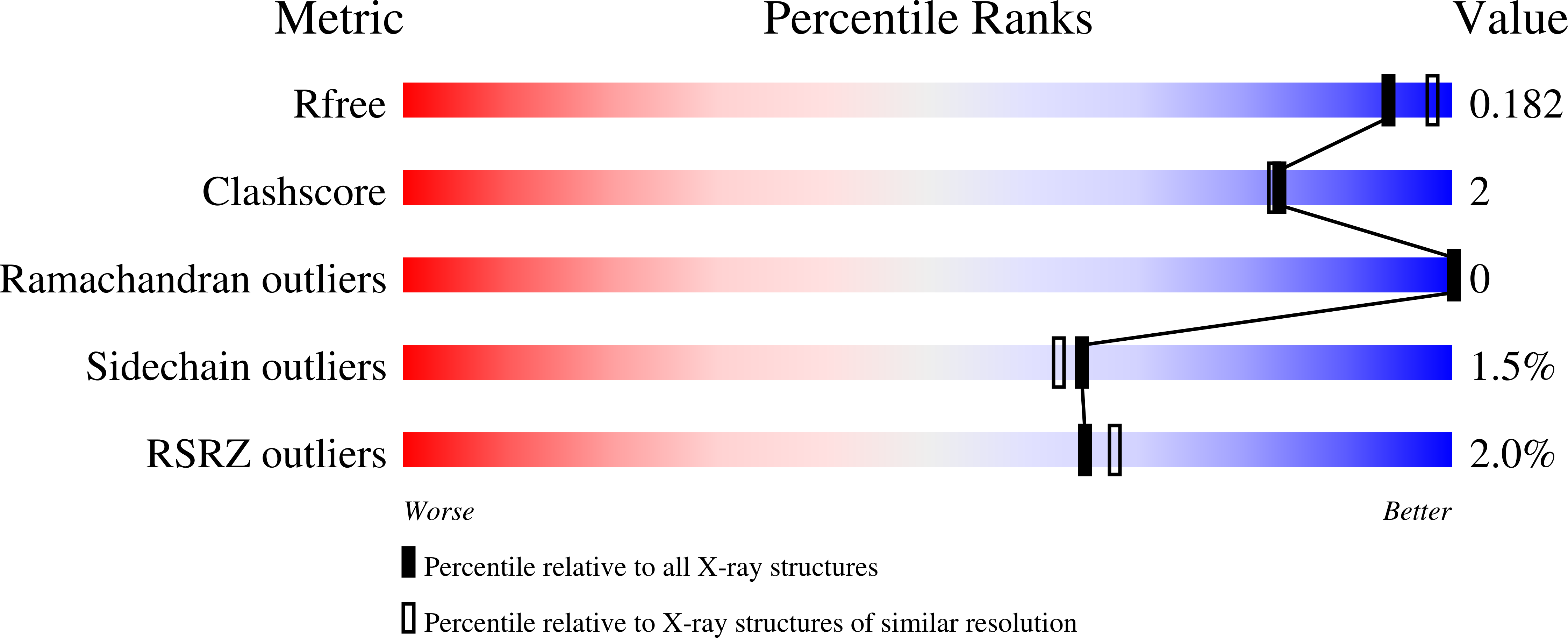
Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes.
Carpena, X., Wiseman, B., Deemagarn, T., Herguedas, B., Ivancich, A., Singh, R., Loewen, P.C., Fita, I.(2006) Biochemistry 45: 5171-5179
- PubMed: 16618106
- DOI: https://doi.org/10.1021/bi060017f
- Primary Citation of Related Structures:
5SX3, 5SX6, 5SX7 - PubMed Abstract:
Crystals of Burkholderia pseudomallei KatG retain their ability to diffract X-rays at high resolution after adjustment of the pH from 5.6 to 4.5, 6.5, 7.5, and 8.5, providing a unique view of the effect of pH on protein structure. One significant pH-sensitive change lies in the appearance of a perhydroxy group attached to the indole nitrogen of the active site Trp111 above pH 7, similar to a modification originally observed in the Ser324Thr variant of the enzyme at pH 5.6. The modification forms rapidly from molecular oxygen in the buffer with 100% occupancy after one minute of soaking of the crystal at room temperature and pH 8.5. The low temperature (4 K) ferric EPR spectra of the resting enzyme, being very sensitive to changes in the heme iron microenvironment, confirm the presence of the modification above pH 7 in native enzyme and variants lacking Arg426 or Met264 and its absence in variants lacking Trp111 or Tyr238. The indole-perhydroxy group is very likely the reactive intermediate of molecular oxygen in the NADH oxidase reaction, and Arg426 is required for its reduction. The second significant pH-sensitive change involves the buried side chain of Arg426 that changes from one predominant conformation at low pH to a second at high pH. The pH profiles of the peroxidase, catalase, and NADH oxidase reactions can be correlated with the distribution of Arg426 conformations. Other pH-induced structural changes include a number of surface-situated side chains, but there is only one change involving a displacement of main chain atoms triggered by the protonation of His53 in a deep pocket in the vicinity of the molecular 2-fold axis.
Organizational Affiliation:
Departament de Biologia Estructural (IBMB-CSIC), Parc Científic de Barcelona, Josep Samitier 1-5, 08028 Barcelona, Spain.